Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
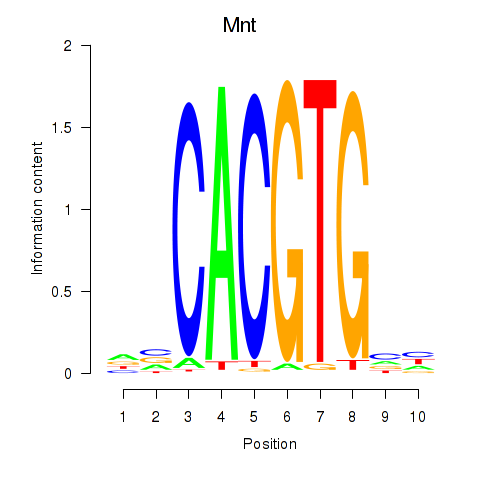
Results for Mnt
Z-value: 1.17
Transcription factors associated with Mnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mnt
|
ENSMUSG00000000282.13 | max binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnt | mm39_v1_chr11_+_74721733_74721746 | 0.68 | 5.7e-06 | Click! |
Activity profile of Mnt motif
Sorted Z-values of Mnt motif
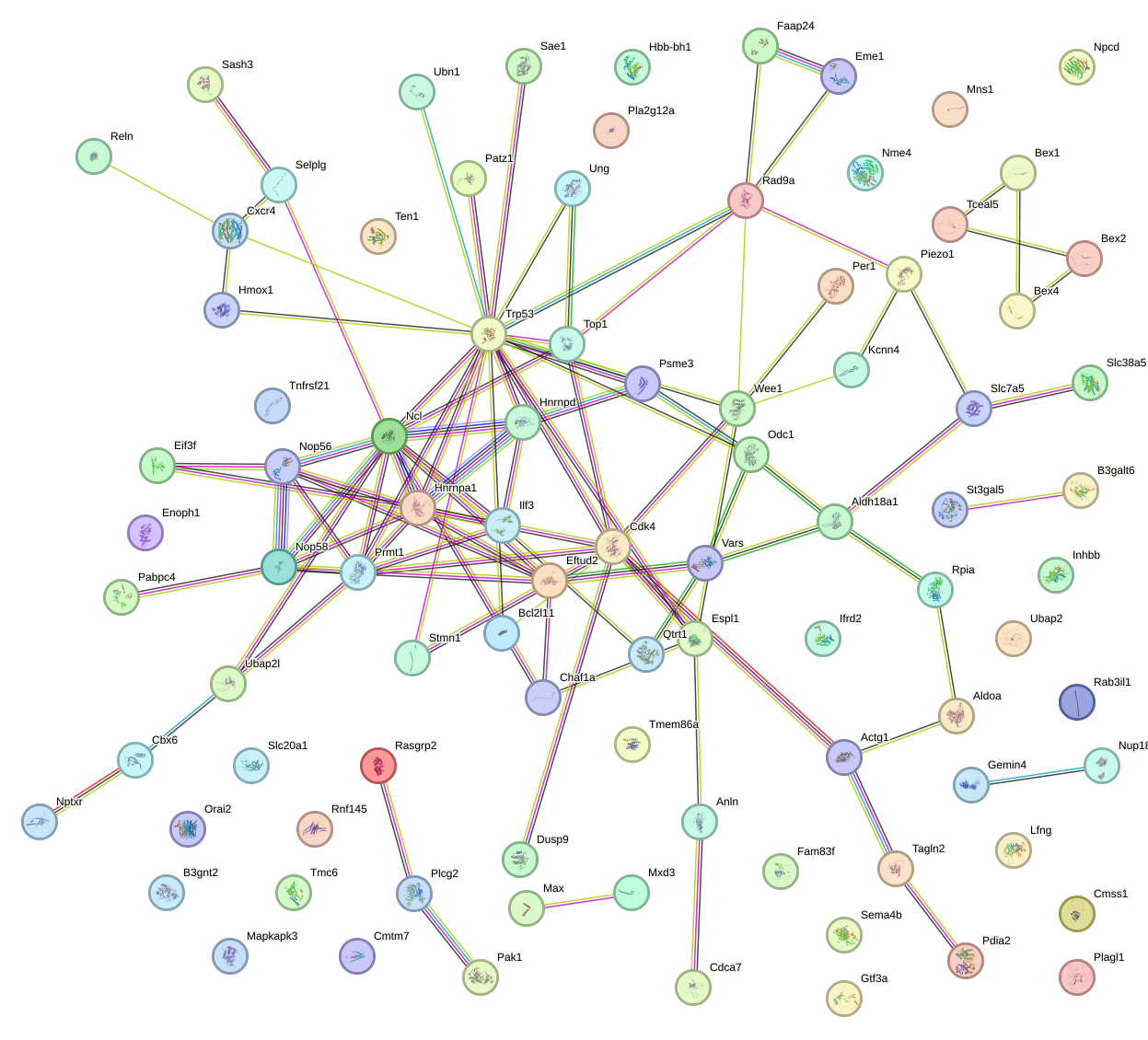
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnt
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 2.7 | 13.7 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 2.0 | 11.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.9 | 13.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 1.8 | 5.5 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 1.7 | 10.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 1.6 | 4.7 | GO:0006788 | heme oxidation(GO:0006788) |
| 1.5 | 4.5 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 1.5 | 4.4 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 1.3 | 5.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 1.2 | 4.6 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 1.0 | 3.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.0 | 6.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 1.0 | 9.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 1.0 | 5.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 1.0 | 3.9 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.0 | 4.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.0 | 5.8 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.9 | 16.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.9 | 2.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.9 | 3.7 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.9 | 4.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.9 | 2.6 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.9 | 1.7 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.8 | 5.9 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.8 | 4.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.8 | 3.3 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.8 | 2.4 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.8 | 14.7 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.7 | 2.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.7 | 3.5 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.7 | 2.7 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.7 | 2.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.7 | 4.6 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.6 | 21.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.6 | 2.6 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.6 | 6.8 | GO:1901249 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.6 | 0.6 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.6 | 2.4 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.6 | 4.0 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.6 | 2.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.6 | 7.8 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.5 | 3.7 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.5 | 2.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.5 | 1.5 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.5 | 2.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.5 | 2.0 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.5 | 2.0 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.5 | 2.0 | GO:1904008 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.5 | 7.9 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.5 | 1.5 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.5 | 8.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 2.4 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.5 | 1.4 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) |
| 0.5 | 0.9 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.5 | 3.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.4 | 4.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.4 | 0.9 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.4 | 6.7 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.4 | 4.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.4 | 0.8 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.4 | 4.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 12.8 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.4 | 1.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.4 | 4.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.4 | 5.7 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.4 | 1.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 0.4 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.4 | 3.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 2.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 2.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.3 | 2.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 2.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.3 | 7.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.3 | 1.3 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.3 | 0.3 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.3 | 1.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.3 | 2.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 8.8 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.3 | 19.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.3 | 1.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.3 | 1.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 2.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 1.1 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.3 | 6.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.3 | 4.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 0.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 0.7 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.2 | 2.7 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 1.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.7 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) seminal vesicle development(GO:0061107) |
| 0.2 | 5.5 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 | 1.6 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 1.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 1.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 1.1 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.2 | 0.9 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 2.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 1.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 2.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.2 | 1.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 4.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 2.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 1.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 1.0 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 1.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.2 | 1.7 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 0.6 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.2 | 0.4 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.2 | 0.9 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.2 | 10.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 4.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 3.7 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 | 0.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 1.3 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.2 | 0.7 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.2 | 1.7 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.2 | 0.7 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 0.9 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.2 | 2.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 0.5 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.2 | 1.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 1.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 1.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 1.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 1.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 1.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 1.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 0.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.2 | 3.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 4.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 2.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 1.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.6 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 3.8 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 2.0 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.9 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 1.6 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.3 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.1 | 1.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 2.4 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.1 | 6.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.9 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 12.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.4 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 1.1 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 1.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.7 | GO:0055099 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.5 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.1 | 4.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 3.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 3.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.6 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 1.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 4.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 1.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 3.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 7.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.4 | GO:0032986 | protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.4 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.6 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 6.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.5 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.6 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.3 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.9 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 0.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.6 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.3 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 1.0 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.5 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.4 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 4.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 0.3 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.1 | 2.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 1.9 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.1 | 0.7 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 0.3 | GO:0061215 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.2 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.1 | 1.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.5 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.1 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.1 | 1.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.1 | 0.3 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.1 | 2.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 2.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 3.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 5.4 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.1 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.4 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 3.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 2.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 1.1 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 2.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.8 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.7 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 2.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 1.3 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.9 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 1.6 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.6 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.6 | GO:0070875 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.5 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.4 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 1.1 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 1.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:1905077 | regulation of interleukin-17 secretion(GO:1905076) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 2.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 2.6 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.2 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.1 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 2.6 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 2.8 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) |
| 0.0 | 1.1 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.7 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.2 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.6 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 1.6 | 14.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 1.5 | 4.6 | GO:1990879 | CST complex(GO:1990879) |
| 1.5 | 4.4 | GO:0097144 | BAX complex(GO:0097144) |
| 1.4 | 7.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.4 | 10.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.3 | 8.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 1.2 | 8.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.2 | 5.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 1.1 | 13.7 | GO:0034709 | methylosome(GO:0034709) |
| 1.0 | 4.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.0 | 5.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.8 | 2.5 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.8 | 3.3 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.7 | 2.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.6 | 3.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.6 | 4.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.6 | 4.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.6 | 4.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.5 | 1.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.5 | 3.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.5 | 5.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 10.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 1.5 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.4 | 3.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.4 | 1.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.3 | 1.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 2.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 1.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 2.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 3.6 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.3 | 0.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.3 | 1.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 1.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 2.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 0.8 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 1.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.3 | 1.6 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 0.3 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.3 | 2.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 1.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 3.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 2.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 1.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 1.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 6.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 2.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 4.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 1.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 2.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 3.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 8.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 1.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 14.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.2 | 2.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 0.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 6.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 3.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 11.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 2.0 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 2.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 1.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 5.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 2.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 3.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 8.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 2.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 1.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 5.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 2.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 3.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.0 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 4.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 4.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 17.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 5.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.8 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 4.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 8.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 5.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 5.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 2.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 5.0 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 3.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 2.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 23.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 29.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 2.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 3.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 3.6 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 13.7 | GO:0030519 | snoRNP binding(GO:0030519) |
| 3.3 | 10.0 | GO:0019202 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 3.2 | 9.6 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 2.4 | 7.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 1.7 | 10.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 1.6 | 4.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.3 | 6.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.2 | 3.7 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 1.2 | 16.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 1.2 | 5.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.2 | 5.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 1.1 | 4.5 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.9 | 2.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.9 | 2.8 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.9 | 2.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.9 | 3.5 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.9 | 7.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.8 | 2.4 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.7 | 2.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.7 | 2.7 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.7 | 4.6 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.6 | 6.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.6 | 11.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.6 | 2.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.6 | 5.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.6 | 6.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 2.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.6 | 1.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.6 | 3.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.5 | 5.9 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.5 | 5.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 3.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.5 | 5.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 7.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.5 | 2.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.5 | 2.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.5 | 22.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.5 | 1.4 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.4 | 3.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.4 | 11.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 4.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 4.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 2.4 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 4.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 5.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.4 | 3.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 3.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.3 | 1.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 2.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 3.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 3.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.3 | 3.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 0.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 17.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 1.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 1.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.3 | 4.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 1.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 2.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 4.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 1.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 1.7 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 4.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 1.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 2.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 8.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 0.8 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.3 | 0.8 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.3 | 0.8 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 1.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 3.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 1.0 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 1.0 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.2 | 0.7 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.2 | 0.7 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 4.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 3.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 19.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 0.4 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 1.4 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 4.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 1.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 4.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 7.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 2.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 0.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 2.0 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.2 | 1.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 3.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 3.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 8.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 4.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.4 | GO:0086077 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.3 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 2.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 4.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 1.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.5 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 5.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 3.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 5.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 5.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 1.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 5.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 5.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 2.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 11.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 1.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 2.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 9.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 2.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.7 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 1.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 4.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 2.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 16.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 4.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 2.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 2.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 2.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 5.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 2.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 2.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 7.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 4.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 6.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 3.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 14.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 7.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.4 | 9.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 8.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 7.8 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 21.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 12.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 2.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 5.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 5.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 10.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 4.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 2.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 4.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 13.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 5.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 4.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 3.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 3.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 4.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 34.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 5.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 7.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 10.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 10.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 7.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.3 | 3.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 18.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 1.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.3 | 5.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 6.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 3.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 8.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 3.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 10.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 1.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 5.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 6.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 28.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 6.6 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.2 | 5.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 8.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 3.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 7.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 4.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.8 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 15.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 3.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.9 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.1 | 2.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 3.2 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 2.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 4.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.2 | REACTOME S PHASE | Genes involved in S Phase |
| 0.1 | 2.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.9 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 4.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 6.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 4.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 5.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 2.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 3.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 3.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |