Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Mnx1_Lhx6_Lmx1a
Z-value: 0.80
Transcription factors associated with Mnx1_Lhx6_Lmx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
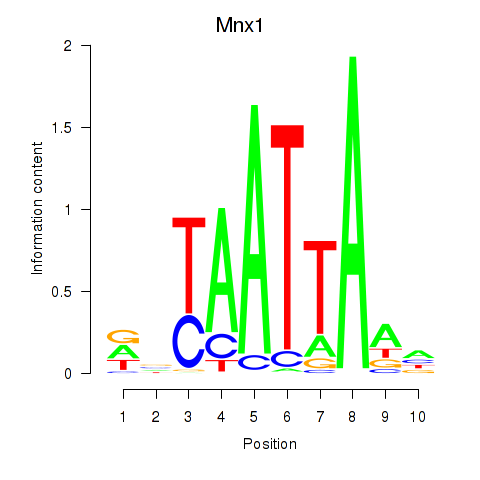
Mnx1
|
ENSMUSG00000001566.10 | motor neuron and pancreas homeobox 1 |
|
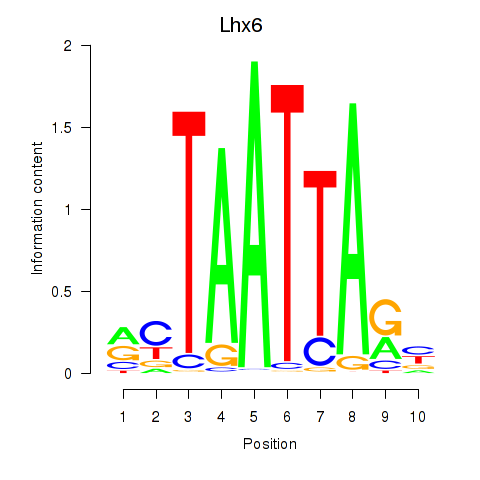
Lhx6
|
ENSMUSG00000026890.20 | LIM homeobox protein 6 |
|
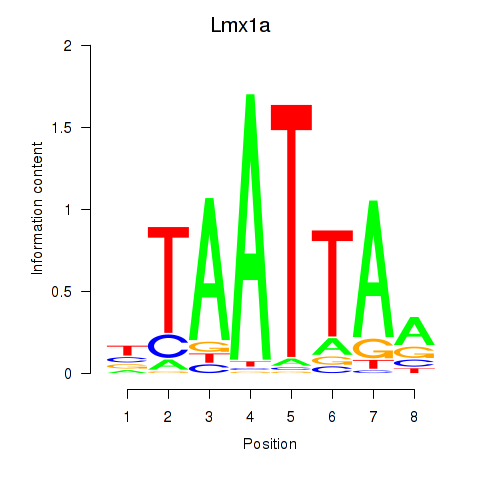
Lmx1a
|
ENSMUSG00000026686.15 | LIM homeobox transcription factor 1 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lmx1a | mm39_v1_chr1_+_167516771_167516806 | 0.39 | 1.7e-02 | Click! |
| Lhx6 | mm39_v1_chr2_-_35994072_35994085 | 0.21 | 2.1e-01 | Click! |
| Mnx1 | mm39_v1_chr5_-_29683468_29683595 | 0.16 | 3.4e-01 | Click! |
Activity profile of Mnx1_Lhx6_Lmx1a motif
Sorted Z-values of Mnx1_Lhx6_Lmx1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnx1_Lhx6_Lmx1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.8 | 5.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 1.4 | 4.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.3 | 10.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.1 | 3.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.9 | 2.6 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.8 | 2.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.7 | 2.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.7 | 2.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.7 | 2.1 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.7 | 2.7 | GO:0090309 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.7 | 2.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.6 | 1.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 1.6 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.5 | 2.6 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.5 | 5.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 2.8 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.5 | 1.8 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.4 | 1.8 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.4 | 1.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.4 | 1.4 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.3 | 2.7 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 4.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.3 | 2.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 2.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 0.8 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.3 | 0.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.2 | 1.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 2.0 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.2 | 2.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 2.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 0.6 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.2 | 0.9 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 3.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 0.4 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.2 | 0.8 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.2 | 0.7 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.2 | 1.6 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 0.7 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.5 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.2 | 1.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.2 | 3.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.8 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.9 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 2.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.4 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.4 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.4 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.6 | GO:0002358 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.1 | 3.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.5 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 2.0 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.3 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.4 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 | 1.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.6 | GO:0060686 | esophagus smooth muscle contraction(GO:0014846) negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.3 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.1 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.6 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 4.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.8 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 2.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.8 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 4.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.6 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 2.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 1.0 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 2.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 1.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.5 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.8 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 1.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 1.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 2.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 1.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0048021 | regulation of melanin biosynthetic process(GO:0048021) regulation of secondary metabolite biosynthetic process(GO:1900376) |
| 0.0 | 0.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.5 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 1.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 3.8 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.1 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.0 | 0.8 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0061296 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.0 | 1.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 2.6 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.5 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.3 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.0 | 0.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 1.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) optic cup structural organization(GO:0003409) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.6 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 2.5 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 6.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 3.5 | GO:0042254 | ribosome biogenesis(GO:0042254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.9 | 2.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 5.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.7 | 3.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.7 | 4.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 3.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.0 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.3 | 1.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 2.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 2.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 2.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 4.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 5.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 2.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 2.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 1.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 9.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 2.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 3.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 2.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.0 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 5.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.7 | 4.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.5 | 2.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 1.6 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.4 | 2.8 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.3 | 2.7 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 1.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 2.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.3 | 4.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 10.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 2.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.7 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.2 | 1.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 3.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 2.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.8 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 5.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 1.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 2.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 2.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 2.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 7.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.7 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.4 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 1.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 3.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 1.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 3.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 2.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 2.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 2.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 1.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 5.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 2.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 3.4 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 3.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 6.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 3.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.7 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 5.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 11.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 3.4 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 3.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 3.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 3.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.4 | 10.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 3.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 2.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 8.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 7.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 2.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |