Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
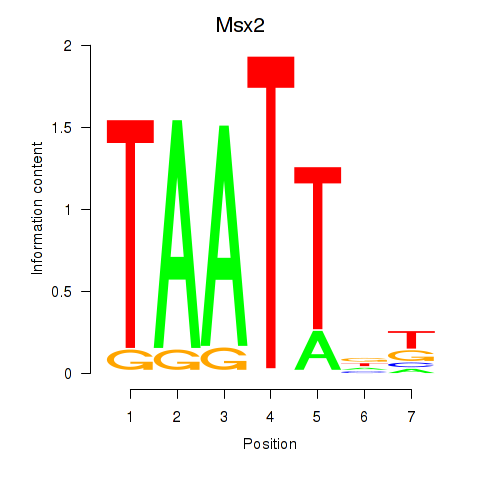
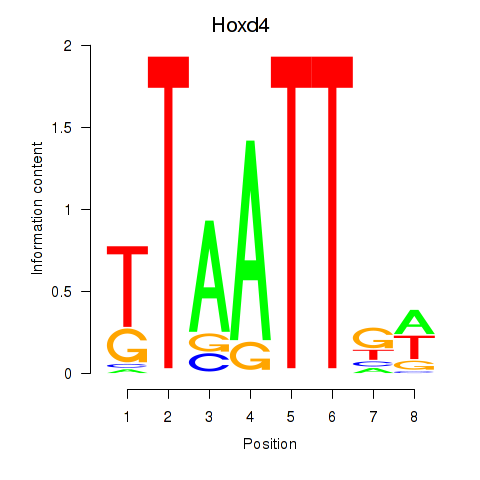
Results for Msx2_Hoxd4
Z-value: 0.68
Transcription factors associated with Msx2_Hoxd4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx2
|
ENSMUSG00000021469.10 | msh homeobox 2 |
|
Hoxd4
|
ENSMUSG00000101174.9 | homeobox D4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx2 | mm39_v1_chr13_-_53627110_53627110 | 0.39 | 1.7e-02 | Click! |
Activity profile of Msx2_Hoxd4 motif
Sorted Z-values of Msx2_Hoxd4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_4022537 | 3.80 |
ENSMUST00000177457.8
|
Frmd4a
|
FERM domain containing 4A |
| chr1_-_132318039 | 3.75 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr15_+_6673167 | 3.32 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr5_+_117378510 | 2.70 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chrX_+_135145813 | 2.55 |
ENSMUST00000048687.11
|
Tceal9
|
transcription elongation factor A like 9 |
| chr18_-_43610829 | 2.44 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr3_-_14843512 | 2.26 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr1_+_45834645 | 2.17 |
ENSMUST00000147308.2
|
Wdr75
|
WD repeat domain 75 |
| chrX_+_55500170 | 1.89 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr6_-_136918885 | 1.75 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chrX_+_149330371 | 1.61 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr18_+_34973605 | 1.58 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr7_-_115459082 | 1.56 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr5_-_62923463 | 1.56 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr19_-_46033353 | 1.54 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr7_-_120673761 | 1.47 |
ENSMUST00000047194.4
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr7_+_89814713 | 1.39 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_+_45433899 | 1.35 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr6_-_87510200 | 1.29 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr6_-_36787096 | 1.08 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr17_+_46471950 | 1.08 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr1_-_149836974 | 1.07 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr7_-_73191484 | 1.05 |
ENSMUST00000197642.2
ENSMUST00000026895.14 ENSMUST00000169922.9 |
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr7_-_45570828 | 1.00 |
ENSMUST00000038876.13
|
Emp3
|
epithelial membrane protein 3 |
| chr9_-_114219685 | 0.96 |
ENSMUST00000084881.5
|
Crtap
|
cartilage associated protein |
| chr7_-_55669702 | 0.96 |
ENSMUST00000052204.6
|
Nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) |
| chr11_-_79421397 | 0.89 |
ENSMUST00000103236.4
ENSMUST00000170799.8 ENSMUST00000170422.4 |
Evi2a
Evi2
|
ecotropic viral integration site 2a ecotropic viral integration site 2 |
| chr1_-_165535654 | 0.87 |
ENSMUST00000097474.9
|
Rcsd1
|
RCSD domain containing 1 |
| chr7_-_45570254 | 0.85 |
ENSMUST00000164119.3
|
Emp3
|
epithelial membrane protein 3 |
| chr7_-_45570538 | 0.83 |
ENSMUST00000210297.2
|
Emp3
|
epithelial membrane protein 3 |
| chr18_+_4920513 | 0.83 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr11_+_67061908 | 0.83 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr11_+_67061837 | 0.82 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chrX_+_100683662 | 0.82 |
ENSMUST00000119299.8
ENSMUST00000044475.5 |
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) |
| chr5_-_23821523 | 0.81 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr6_+_68233361 | 0.79 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chrX_+_158491589 | 0.78 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_-_115423934 | 0.77 |
ENSMUST00000169129.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr14_+_79753055 | 0.77 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr2_-_84255602 | 0.75 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr7_-_45570674 | 0.75 |
ENSMUST00000210939.2
|
Emp3
|
epithelial membrane protein 3 |
| chr9_-_107648144 | 0.73 |
ENSMUST00000183248.3
ENSMUST00000182022.8 ENSMUST00000035199.13 ENSMUST00000182659.8 |
Rbm5
|
RNA binding motif protein 5 |
| chr6_-_129449739 | 0.72 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr2_-_163259012 | 0.71 |
ENSMUST00000127038.2
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chrX_+_158086253 | 0.71 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr6_+_136495784 | 0.69 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr18_+_7905440 | 0.67 |
ENSMUST00000170854.2
|
Wac
|
WW domain containing adaptor with coiled-coil |
| chr2_-_45002902 | 0.66 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_+_83142902 | 0.64 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr1_-_80255156 | 0.62 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr2_-_73284262 | 0.58 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_128079543 | 0.58 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1 |
| chr11_+_98689479 | 0.58 |
ENSMUST00000037930.13
|
Msl1
|
male specific lethal 1 |
| chr5_+_65505657 | 0.57 |
ENSMUST00000031096.11
|
Klb
|
klotho beta |
| chr13_-_100753419 | 0.57 |
ENSMUST00000168772.2
ENSMUST00000163163.9 ENSMUST00000022137.14 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr9_-_56151334 | 0.56 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr6_+_29361408 | 0.55 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr17_+_71923210 | 0.53 |
ENSMUST00000047086.10
|
Wdr43
|
WD repeat domain 43 |
| chr4_+_3940747 | 0.50 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr2_+_85809620 | 0.50 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr2_-_59955995 | 0.49 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr3_+_40755211 | 0.49 |
ENSMUST00000204473.2
|
Plk4
|
polo like kinase 4 |
| chr10_+_5543769 | 0.48 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr6_+_34722887 | 0.44 |
ENSMUST00000123823.8
ENSMUST00000136907.8 |
Cald1
|
caldesmon 1 |
| chr3_+_84859453 | 0.43 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr9_-_106666329 | 0.43 |
ENSMUST00000046502.7
|
Rad54l2
|
RAD54 like 2 (S. cerevisiae) |
| chr5_-_72325482 | 0.42 |
ENSMUST00000196241.2
ENSMUST00000013693.11 |
Commd8
|
COMM domain containing 8 |
| chr11_-_74615496 | 0.42 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr17_-_37523969 | 0.39 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr6_+_136495818 | 0.38 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr13_+_51562675 | 0.37 |
ENSMUST00000087978.5
|
S1pr3
|
sphingosine-1-phosphate receptor 3 |
| chr6_-_69584812 | 0.37 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr10_+_128139191 | 0.37 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr3_+_32490300 | 0.37 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr5_-_108022900 | 0.36 |
ENSMUST00000138111.8
ENSMUST00000112642.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr7_-_84328553 | 0.36 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr19_-_24178000 | 0.34 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr13_-_76091931 | 0.34 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr18_+_23885390 | 0.34 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_86986541 | 0.33 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr6_+_34722926 | 0.33 |
ENSMUST00000126181.8
|
Cald1
|
caldesmon 1 |
| chr15_+_41694317 | 0.31 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr13_-_100753181 | 0.31 |
ENSMUST00000225754.2
|
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr1_+_179788675 | 0.30 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_+_179936757 | 0.30 |
ENSMUST00000143176.8
ENSMUST00000135056.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_+_34723304 | 0.29 |
ENSMUST00000142716.3
|
Cald1
|
caldesmon 1 |
| chr3_-_126918491 | 0.28 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain |
| chr18_-_33346885 | 0.28 |
ENSMUST00000025236.9
|
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr13_+_49761506 | 0.28 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr4_+_109092610 | 0.27 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr3_+_132335575 | 0.27 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr8_+_107757847 | 0.27 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr10_+_127257077 | 0.26 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr5_-_123127346 | 0.26 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr2_-_45000389 | 0.25 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_+_87404246 | 0.25 |
ENSMUST00000213315.2
ENSMUST00000214773.2 |
Olfr1129
|
olfactory receptor 1129 |
| chr14_+_27598021 | 0.25 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr12_-_113733922 | 0.25 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr6_+_29853745 | 0.25 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr10_-_129107354 | 0.25 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr10_+_127256736 | 0.25 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr18_+_44237577 | 0.25 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr13_+_19398273 | 0.24 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr6_+_15196950 | 0.24 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr10_+_127256993 | 0.24 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr3_+_7494108 | 0.23 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr18_-_33346819 | 0.23 |
ENSMUST00000119991.8
ENSMUST00000118990.2 |
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr7_+_122723365 | 0.23 |
ENSMUST00000205514.2
ENSMUST00000094053.7 |
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr6_+_149226891 | 0.22 |
ENSMUST00000189837.2
|
Resf1
|
retroelement silencing factor 1 |
| chr9_+_38788422 | 0.22 |
ENSMUST00000078289.3
|
Olfr926
|
olfactory receptor 926 |
| chr8_-_26609153 | 0.22 |
ENSMUST00000037182.14
|
Hook3
|
hook microtubule tethering protein 3 |
| chr1_+_157353696 | 0.18 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr14_-_52273600 | 0.18 |
ENSMUST00000214342.2
|
Olfr221
|
olfactory receptor 221 |
| chr8_+_31601837 | 0.18 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr4_+_101365144 | 0.18 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr19_+_31846154 | 0.16 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr10_-_63039709 | 0.16 |
ENSMUST00000095580.3
|
Mypn
|
myopalladin |
| chr9_+_74959259 | 0.16 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr4_-_43710231 | 0.16 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr8_-_49296915 | 0.15 |
ENSMUST00000211812.2
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr18_+_44237474 | 0.15 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr17_-_29226700 | 0.15 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38 |
| chr5_-_102217770 | 0.15 |
ENSMUST00000053177.14
ENSMUST00000174698.2 |
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr6_-_30936013 | 0.15 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr10_-_23977810 | 0.15 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr2_-_88519531 | 0.15 |
ENSMUST00000213545.2
|
Olfr1195
|
olfactory receptor 1195 |
| chr14_+_8283087 | 0.14 |
ENSMUST00000206298.3
ENSMUST00000216079.2 |
Olfr720
|
olfactory receptor 720 |
| chr19_-_55229668 | 0.14 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr19_-_47680528 | 0.13 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr4_+_101365052 | 0.13 |
ENSMUST00000038207.12
|
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr2_+_36342599 | 0.13 |
ENSMUST00000072854.2
|
Olfr340
|
olfactory receptor 340 |
| chr6_+_83985684 | 0.13 |
ENSMUST00000203803.3
ENSMUST00000204591.3 ENSMUST00000113823.8 ENSMUST00000153860.4 |
Dysf
|
dysferlin |
| chr1_-_156766351 | 0.13 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr4_+_109092459 | 0.13 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr1_-_155293141 | 0.12 |
ENSMUST00000111775.8
ENSMUST00000111774.2 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr11_+_76795346 | 0.12 |
ENSMUST00000072633.4
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr13_+_21546990 | 0.11 |
ENSMUST00000225545.2
ENSMUST00000053293.14 |
Zscan12
|
zinc finger and SCAN domain containing 12 |
| chr6_+_57180275 | 0.11 |
ENSMUST00000226892.2
ENSMUST00000227421.2 |
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr2_+_83554741 | 0.11 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr18_+_37453427 | 0.11 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr5_+_107655487 | 0.10 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr11_-_73742280 | 0.10 |
ENSMUST00000213365.2
|
Olfr393
|
olfactory receptor 393 |
| chr1_+_179788037 | 0.10 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_-_156766381 | 0.10 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr6_-_129600798 | 0.10 |
ENSMUST00000095412.10
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr12_-_113896002 | 0.09 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr11_-_99265721 | 0.09 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr10_+_23952398 | 0.09 |
ENSMUST00000051133.6
|
Taar8a
|
trace amine-associated receptor 8A |
| chr16_-_59092995 | 0.09 |
ENSMUST00000216834.2
|
Olfr201
|
olfactory receptor 201 |
| chr3_-_144275897 | 0.09 |
ENSMUST00000043325.9
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr1_+_174192688 | 0.08 |
ENSMUST00000217962.2
ENSMUST00000220394.2 |
Olfr417
|
olfactory receptor 417 |
| chr4_+_13784749 | 0.08 |
ENSMUST00000098256.4
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr8_-_3675274 | 0.08 |
ENSMUST00000004749.7
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr9_+_77959206 | 0.08 |
ENSMUST00000024104.9
|
Gcm1
|
glial cells missing homolog 1 |
| chr14_-_36641270 | 0.07 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr6_-_129600812 | 0.07 |
ENSMUST00000168919.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr7_+_107679062 | 0.06 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr2_+_83554868 | 0.06 |
ENSMUST00000111740.9
|
Itgav
|
integrin alpha V |
| chr4_-_3872105 | 0.06 |
ENSMUST00000105158.2
|
Mos
|
Moloney sarcoma oncogene |
| chr4_-_82768958 | 0.06 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr14_+_53599724 | 0.05 |
ENSMUST00000196105.2
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr3_+_67799510 | 0.05 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr14_+_8282925 | 0.05 |
ENSMUST00000217642.2
|
Olfr720
|
olfactory receptor 720 |
| chr15_-_101422054 | 0.04 |
ENSMUST00000230067.3
|
Gm49425
|
predicted gene, 49425 |
| chr17_-_90395771 | 0.04 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr5_-_121641461 | 0.04 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr10_+_89906956 | 0.04 |
ENSMUST00000183109.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr17_-_21006419 | 0.04 |
ENSMUST00000233605.2
ENSMUST00000232812.2 ENSMUST00000233186.2 ENSMUST00000233164.2 |
Vmn1r228
|
vomeronasal 1 receptor 228 |
| chr6_-_13871475 | 0.04 |
ENSMUST00000139231.2
|
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr13_+_49697919 | 0.03 |
ENSMUST00000177948.2
ENSMUST00000021820.14 |
Aspn
|
asporin |
| chr14_-_4506874 | 0.03 |
ENSMUST00000224934.2
|
Thrb
|
thyroid hormone receptor beta |
| chr1_+_106866678 | 0.03 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr10_+_115405891 | 0.02 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr2_+_109522781 | 0.02 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr15_+_79999643 | 0.02 |
ENSMUST00000135727.2
|
Syngr1
|
synaptogyrin 1 |
| chr1_-_54233207 | 0.02 |
ENSMUST00000120904.8
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_+_20742115 | 0.01 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr19_+_26727111 | 0.01 |
ENSMUST00000175842.4
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_+_129601351 | 0.01 |
ENSMUST00000203236.3
|
Olfr808
|
olfactory receptor 808 |
| chr4_+_80752535 | 0.01 |
ENSMUST00000102831.8
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr7_-_10011933 | 0.01 |
ENSMUST00000227719.2
ENSMUST00000228622.2 ENSMUST00000228086.2 |
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr2_+_14828903 | 0.00 |
ENSMUST00000193800.6
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr14_+_50360643 | 0.00 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr14_-_50586329 | 0.00 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr3_+_106020545 | 0.00 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr3_+_14545751 | 0.00 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx2_Hoxd4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 1.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.3 | 2.7 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.3 | 0.8 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 | 1.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.3 | 1.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.2 | 1.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 1.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 2.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 1.7 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 0.7 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 1.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 0.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.7 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.4 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 3.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.5 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 1.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.9 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.1 | 2.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.6 | GO:0071630 | trophectodermal cellular morphogenesis(GO:0001831) nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 3.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.3 | GO:0021592 | fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.5 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.2 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.1 | 3.7 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 2.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 1.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 1.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 3.1 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.4 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.9 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.1 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 2.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.1 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 1.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 4.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.3 | 0.8 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.3 | 1.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 0.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 2.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 1.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.2 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.0 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 1.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 4.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.4 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 4.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |