Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
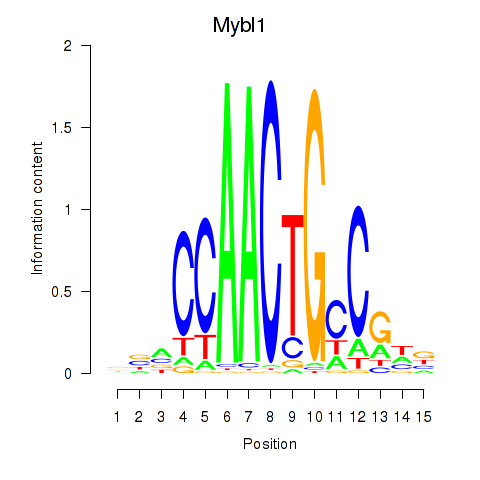
Results for Mybl1
Z-value: 2.25
Transcription factors associated with Mybl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl1
|
ENSMUSG00000025912.17 | myeloblastosis oncogene-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl1 | mm39_v1_chr1_-_9770434_9770554 | 0.39 | 1.8e-02 | Click! |
Activity profile of Mybl1 motif
Sorted Z-values of Mybl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87684299 | 22.95 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr14_-_56339915 | 22.73 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr4_-_132260799 | 21.61 |
ENSMUST00000152993.8
ENSMUST00000067496.7 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr19_-_11618165 | 20.39 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr4_-_46413484 | 20.04 |
ENSMUST00000071096.3
|
Hemgn
|
hemogen |
| chr19_-_11618192 | 18.71 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr11_+_87684548 | 16.21 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr9_-_109678685 | 15.81 |
ENSMUST00000112022.5
|
Camp
|
cathelicidin antimicrobial peptide |
| chr11_+_32233511 | 14.36 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr2_+_174292471 | 14.14 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr11_+_32246489 | 14.12 |
ENSMUST00000093207.4
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr6_+_124806506 | 12.31 |
ENSMUST00000150120.8
|
Cdca3
|
cell division cycle associated 3 |
| chr6_+_124806541 | 12.31 |
ENSMUST00000024270.14
|
Cdca3
|
cell division cycle associated 3 |
| chr11_+_58808830 | 11.93 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr17_+_48554786 | 10.96 |
ENSMUST00000048065.6
|
Trem3
|
triggering receptor expressed on myeloid cells 3 |
| chr3_+_10077608 | 10.75 |
ENSMUST00000029046.9
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr3_-_36626101 | 10.54 |
ENSMUST00000029270.10
|
Ccna2
|
cyclin A2 |
| chr14_+_46997984 | 10.31 |
ENSMUST00000067426.6
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr7_-_126736979 | 10.17 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr1_-_133681419 | 10.13 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr2_+_118644717 | 10.02 |
ENSMUST00000028803.14
ENSMUST00000126045.8 |
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr8_+_84682136 | 9.45 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr1_+_134110142 | 9.29 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chrX_-_101200670 | 9.28 |
ENSMUST00000056904.3
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr4_-_119047146 | 9.24 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr2_+_118644475 | 9.03 |
ENSMUST00000134661.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr2_+_118644675 | 8.69 |
ENSMUST00000110842.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding |
| chr3_-_68952057 | 8.55 |
ENSMUST00000107802.8
|
Trim59
|
tripartite motif-containing 59 |
| chr4_+_120523758 | 8.44 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chrX_+_135171049 | 8.32 |
ENSMUST00000113112.2
|
Bex3
|
brain expressed X-linked 3 |
| chr9_+_56344700 | 8.24 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr11_+_7147779 | 8.23 |
ENSMUST00000020704.8
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr15_+_73596602 | 8.06 |
ENSMUST00000230177.2
ENSMUST00000163582.9 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr18_-_35782412 | 7.96 |
ENSMUST00000025211.6
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr7_+_100142544 | 7.90 |
ENSMUST00000126534.8
ENSMUST00000207748.2 |
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_-_69496655 | 7.83 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr8_+_84442133 | 7.78 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a |
| chr8_+_84441806 | 7.64 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a |
| chr6_+_30723540 | 7.56 |
ENSMUST00000141130.2
ENSMUST00000115127.8 |
Mest
|
mesoderm specific transcript |
| chr15_-_36609812 | 7.52 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr9_+_96140781 | 7.44 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr2_-_150510116 | 7.14 |
ENSMUST00000028944.4
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr15_-_96540117 | 6.94 |
ENSMUST00000088454.13
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr4_-_43499608 | 6.93 |
ENSMUST00000136005.3
ENSMUST00000054538.13 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr2_+_150751475 | 6.90 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr1_+_134109888 | 6.73 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr14_+_46998004 | 6.70 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chrX_+_135145813 | 6.57 |
ENSMUST00000048687.11
|
Tceal9
|
transcription elongation factor A like 9 |
| chr8_+_85696396 | 6.50 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr1_+_153628598 | 6.44 |
ENSMUST00000182538.3
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr5_-_114961501 | 6.40 |
ENSMUST00000100850.6
|
1500011B03Rik
|
RIKEN cDNA 1500011B03 gene |
| chr10_+_75409282 | 6.40 |
ENSMUST00000006508.10
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr13_-_55677109 | 6.32 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr3_-_68952030 | 6.30 |
ENSMUST00000136512.3
|
Trim59
|
tripartite motif-containing 59 |
| chr19_-_41790458 | 6.28 |
ENSMUST00000026150.15
ENSMUST00000163265.9 ENSMUST00000177495.2 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr15_-_78456898 | 6.27 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr8_+_85696453 | 6.25 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_84441854 | 6.24 |
ENSMUST00000172396.8
|
Ddx39a
|
DEAD box helicase 39a |
| chr5_-_31102829 | 6.23 |
ENSMUST00000031051.8
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr2_-_165242307 | 6.21 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr11_-_72441054 | 6.20 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr4_-_117039809 | 6.18 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr2_-_164198427 | 5.85 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr17_-_79662514 | 5.85 |
ENSMUST00000068958.9
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr14_+_66043281 | 5.82 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr4_-_131695135 | 5.81 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr17_+_28426752 | 5.75 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr17_-_26314461 | 5.73 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr15_+_75027089 | 5.68 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr8_-_106578613 | 5.66 |
ENSMUST00000040776.6
|
Cenpt
|
centromere protein T |
| chr15_-_89310060 | 5.65 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr5_+_123887759 | 5.58 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr2_+_125089110 | 5.55 |
ENSMUST00000082122.14
|
Dut
|
deoxyuridine triphosphatase |
| chr8_-_71725011 | 5.54 |
ENSMUST00000110071.3
|
Haus8
|
4HAUS augmin-like complex, subunit 8 |
| chr19_+_4204605 | 5.49 |
ENSMUST00000061086.9
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr11_+_68792364 | 5.48 |
ENSMUST00000073471.13
ENSMUST00000101014.9 ENSMUST00000128952.8 ENSMUST00000167436.3 |
Rpl26
|
ribosomal protein L26 |
| chr2_+_30331839 | 5.45 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr6_-_128868068 | 5.43 |
ENSMUST00000178918.2
ENSMUST00000160290.8 |
BC035044
|
cDNA sequence BC035044 |
| chr3_+_89638044 | 5.41 |
ENSMUST00000029563.14
ENSMUST00000121094.8 ENSMUST00000118341.6 ENSMUST00000107405.6 |
Adar
|
adenosine deaminase, RNA-specific |
| chr13_+_13612136 | 5.39 |
ENSMUST00000005532.9
|
Nid1
|
nidogen 1 |
| chr14_+_71011744 | 5.26 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr5_+_123214332 | 5.19 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr9_+_64718596 | 5.16 |
ENSMUST00000038890.6
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr17_-_46940346 | 5.16 |
ENSMUST00000044442.10
|
Ptk7
|
PTK7 protein tyrosine kinase 7 |
| chr9_+_72345801 | 5.15 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr4_-_44167509 | 5.08 |
ENSMUST00000098098.9
|
Rnf38
|
ring finger protein 38 |
| chr8_+_85696216 | 5.08 |
ENSMUST00000109734.8
ENSMUST00000005292.15 |
Prdx2
|
peroxiredoxin 2 |
| chr13_-_24945844 | 5.02 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chr5_-_44259010 | 4.97 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr5_-_44259293 | 4.91 |
ENSMUST00000074113.13
|
Prom1
|
prominin 1 |
| chr3_+_30910089 | 4.86 |
ENSMUST00000108261.8
ENSMUST00000108259.8 ENSMUST00000166278.7 ENSMUST00000046748.13 ENSMUST00000194979.6 |
Gpr160
|
G protein-coupled receptor 160 |
| chr1_+_91107725 | 4.84 |
ENSMUST00000188475.7
ENSMUST00000097648.6 |
Ramp1
|
receptor (calcitonin) activity modifying protein 1 |
| chr8_+_118225008 | 4.84 |
ENSMUST00000081232.9
|
Plcg2
|
phospholipase C, gamma 2 |
| chr7_+_142030744 | 4.81 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr1_-_33708876 | 4.76 |
ENSMUST00000027312.11
|
Prim2
|
DNA primase, p58 subunit |
| chr6_-_122778598 | 4.76 |
ENSMUST00000165884.8
|
Slc2a3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr18_+_34757666 | 4.65 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr7_-_45084012 | 4.64 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr5_+_66833434 | 4.57 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr14_+_22069877 | 4.57 |
ENSMUST00000161249.8
ENSMUST00000159777.8 ENSMUST00000162540.2 |
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr14_+_76714350 | 4.54 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr7_-_45083688 | 4.53 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr9_+_21437440 | 4.52 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr7_+_125043806 | 4.48 |
ENSMUST00000033010.9
ENSMUST00000135129.2 |
Kdm8
|
lysine (K)-specific demethylase 8 |
| chr9_+_62765362 | 4.47 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr5_-_129856237 | 4.43 |
ENSMUST00000118268.9
|
Psph
|
phosphoserine phosphatase |
| chr1_-_167112170 | 4.31 |
ENSMUST00000192269.3
|
Uck2
|
uridine-cytidine kinase 2 |
| chr7_+_126461601 | 4.27 |
ENSMUST00000132808.2
|
Hirip3
|
HIRA interacting protein 3 |
| chr17_+_28059129 | 4.17 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr16_+_45430743 | 4.14 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr15_-_99149810 | 4.11 |
ENSMUST00000163506.3
ENSMUST00000229671.2 ENSMUST00000229359.2 ENSMUST00000041190.17 |
Mcrs1
|
microspherule protein 1 |
| chr1_-_71142305 | 4.08 |
ENSMUST00000027393.8
|
Bard1
|
BRCA1 associated RING domain 1 |
| chr5_+_76331727 | 4.05 |
ENSMUST00000031144.14
|
Tmem165
|
transmembrane protein 165 |
| chr8_+_84335176 | 4.01 |
ENSMUST00000212300.2
|
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr14_-_60324265 | 3.99 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr11_-_109364046 | 3.97 |
ENSMUST00000070872.13
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr15_-_82128538 | 3.97 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr5_+_137627431 | 3.94 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr9_+_96140750 | 3.93 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr2_+_121337226 | 3.88 |
ENSMUST00000099473.10
ENSMUST00000110602.9 |
Wdr76
|
WD repeat domain 76 |
| chr7_+_3339077 | 3.86 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_+_3339059 | 3.84 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr2_+_91480513 | 3.82 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr19_-_17333972 | 3.79 |
ENSMUST00000174236.8
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chrX_+_49930311 | 3.77 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr17_+_28547533 | 3.75 |
ENSMUST00000233427.2
ENSMUST00000233937.2 |
Rpl10a
|
ribosomal protein L10A |
| chr14_+_66043515 | 3.74 |
ENSMUST00000139644.2
|
Pbk
|
PDZ binding kinase |
| chr17_+_28059099 | 3.73 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr7_+_46496506 | 3.72 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr7_-_126461000 | 3.71 |
ENSMUST00000106343.3
ENSMUST00000205349.2 ENSMUST00000206349.2 |
Ino80e
|
INO80 complex subunit E |
| chr11_-_120534469 | 3.71 |
ENSMUST00000141254.8
ENSMUST00000170556.8 ENSMUST00000151876.8 ENSMUST00000026133.15 ENSMUST00000139706.2 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr19_-_46917661 | 3.71 |
ENSMUST00000236727.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr17_+_28547548 | 3.70 |
ENSMUST00000233895.2
ENSMUST00000232867.2 |
Rpl10a
|
ribosomal protein L10A |
| chr10_-_79710468 | 3.70 |
ENSMUST00000092325.11
|
Plppr3
|
phospholipid phosphatase related 3 |
| chr7_+_92524460 | 3.68 |
ENSMUST00000076052.8
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr12_-_69837434 | 3.67 |
ENSMUST00000021377.5
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr4_-_49597837 | 3.64 |
ENSMUST00000042750.3
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr8_+_69246581 | 3.63 |
ENSMUST00000034328.13
ENSMUST00000110241.8 ENSMUST00000110242.8 ENSMUST00000070713.8 |
Ints10
|
integrator complex subunit 10 |
| chr15_-_82128888 | 3.62 |
ENSMUST00000089155.6
ENSMUST00000089157.11 |
Cenpm
|
centromere protein M |
| chr7_+_46496929 | 3.62 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr11_-_34674677 | 3.61 |
ENSMUST00000093193.12
ENSMUST00000101365.9 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr2_+_118943274 | 3.61 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr2_+_80469142 | 3.60 |
ENSMUST00000028382.13
ENSMUST00000124377.2 |
Nup35
|
nucleoporin 35 |
| chr7_+_126461117 | 3.59 |
ENSMUST00000037248.10
|
Hirip3
|
HIRA interacting protein 3 |
| chr5_+_33978035 | 3.53 |
ENSMUST00000075812.11
ENSMUST00000114397.9 ENSMUST00000155880.8 |
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr19_-_7688628 | 3.51 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr3_+_5815863 | 3.51 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr11_+_69471185 | 3.49 |
ENSMUST00000171247.8
ENSMUST00000108658.10 ENSMUST00000005371.12 |
Trp53
|
transformation related protein 53 |
| chr15_+_88635852 | 3.47 |
ENSMUST00000041297.15
|
Zbed4
|
zinc finger, BED type containing 4 |
| chr7_+_46496552 | 3.45 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr2_+_122479770 | 3.42 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr17_+_35133435 | 3.41 |
ENSMUST00000007249.15
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr1_+_153627692 | 3.40 |
ENSMUST00000183241.8
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr17_-_28736483 | 3.39 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr19_-_6191440 | 3.39 |
ENSMUST00000025893.7
|
Arl2
|
ADP-ribosylation factor-like 2 |
| chr1_-_60137294 | 3.37 |
ENSMUST00000141417.3
ENSMUST00000122038.8 |
Wdr12
|
WD repeat domain 12 |
| chr2_+_70948267 | 3.36 |
ENSMUST00000028403.3
|
Cybrd1
|
cytochrome b reductase 1 |
| chr1_-_165462020 | 3.33 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chr6_+_129374441 | 3.30 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr10_+_127512933 | 3.29 |
ENSMUST00000118612.8
ENSMUST00000048099.5 |
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr17_+_28547445 | 3.29 |
ENSMUST00000042334.16
|
Rpl10a
|
ribosomal protein L10A |
| chr9_-_30833748 | 3.27 |
ENSMUST00000065112.7
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr11_+_62442502 | 3.24 |
ENSMUST00000136938.2
|
Ubb
|
ubiquitin B |
| chr4_-_44167904 | 3.22 |
ENSMUST00000102934.9
|
Rnf38
|
ring finger protein 38 |
| chr6_+_35229589 | 3.22 |
ENSMUST00000152147.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr11_+_69471219 | 3.19 |
ENSMUST00000108657.4
|
Trp53
|
transformation related protein 53 |
| chr6_+_35229628 | 3.19 |
ENSMUST00000130875.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr16_+_91203123 | 3.17 |
ENSMUST00000023691.12
|
Il10rb
|
interleukin 10 receptor, beta |
| chr7_-_115630282 | 3.16 |
ENSMUST00000206034.2
ENSMUST00000106612.8 |
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr7_+_92524495 | 3.14 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C) |
| chr5_-_33809872 | 3.12 |
ENSMUST00000057551.14
|
Slbp
|
stem-loop binding protein |
| chr4_+_21776261 | 3.12 |
ENSMUST00000065111.15
ENSMUST00000040429.12 ENSMUST00000148304.8 |
Usp45
|
ubiquitin specific petidase 45 |
| chr5_-_114952003 | 3.11 |
ENSMUST00000112160.4
|
1500011B03Rik
|
RIKEN cDNA 1500011B03 gene |
| chr19_+_53131187 | 3.08 |
ENSMUST00000050096.15
ENSMUST00000237832.2 |
Add3
|
adducin 3 (gamma) |
| chr2_-_105229653 | 3.08 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr9_+_64718708 | 3.07 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr9_+_106158549 | 3.03 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chr12_-_99849660 | 3.02 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr7_+_97492124 | 3.00 |
ENSMUST00000033040.12
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr2_+_153583194 | 2.97 |
ENSMUST00000028981.9
|
Mapre1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr11_-_75239084 | 2.93 |
ENSMUST00000000767.6
ENSMUST00000092907.12 |
Rpa1
|
replication protein A1 |
| chr9_-_114811807 | 2.92 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr8_+_36561982 | 2.90 |
ENSMUST00000110492.2
|
Prag1
|
PEAK1 related kinase activating pseudokinase 1 |
| chr19_-_4861536 | 2.89 |
ENSMUST00000179909.8
ENSMUST00000172000.3 ENSMUST00000180008.2 ENSMUST00000113793.4 ENSMUST00000006625.8 |
Gm21992
Rbm14
|
predicted gene 21992 RNA binding motif protein 14 |
| chr14_-_99231754 | 2.89 |
ENSMUST00000081987.5
|
Rpl36a-ps1
|
ribosomal protein L36A, pseudogene 1 |
| chr4_-_44168339 | 2.88 |
ENSMUST00000045793.15
|
Rnf38
|
ring finger protein 38 |
| chr8_+_84334805 | 2.86 |
ENSMUST00000005620.10
|
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr2_-_25162347 | 2.83 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr1_+_180762587 | 2.79 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr13_-_55169100 | 2.78 |
ENSMUST00000148221.8
ENSMUST00000052949.13 |
Hk3
|
hexokinase 3 |
| chr8_-_31658775 | 2.77 |
ENSMUST00000033983.6
|
Mak16
|
MAK16 homolog |
| chr15_+_52575796 | 2.72 |
ENSMUST00000037115.9
|
Med30
|
mediator complex subunit 30 |
| chr14_+_24540745 | 2.69 |
ENSMUST00000112384.10
|
Rps24
|
ribosomal protein S24 |
| chr10_+_110581293 | 2.69 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chr16_+_13981139 | 2.67 |
ENSMUST00000023359.13
ENSMUST00000117958.8 |
Nde1
|
nudE neurodevelopment protein 1 |
| chr15_-_99149794 | 2.67 |
ENSMUST00000229926.2
|
Mcrs1
|
microspherule protein 1 |
| chr10_-_22607136 | 2.67 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr11_+_101215889 | 2.66 |
ENSMUST00000041095.14
ENSMUST00000107264.2 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr11_-_68864666 | 2.65 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr3_-_129625023 | 2.60 |
ENSMUST00000029643.15
|
Gar1
|
GAR1 ribonucleoprotein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 39.2 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 3.6 | 21.6 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 3.4 | 10.2 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 3.4 | 10.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 3.4 | 33.7 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 2.4 | 7.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 2.2 | 6.7 | GO:1904024 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 2.0 | 15.8 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 2.0 | 9.9 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.8 | 5.5 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 1.8 | 5.4 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 1.8 | 8.8 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 1.7 | 6.8 | GO:0002353 | plasma kallikrein-kinin cascade(GO:0002353) |
| 1.6 | 19.2 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 1.6 | 7.8 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 1.6 | 6.2 | GO:0090290 | positive regulation of macrophage fusion(GO:0034241) positive regulation of osteoclast proliferation(GO:0090290) |
| 1.5 | 7.7 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 1.5 | 10.8 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.5 | 9.2 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 1.4 | 4.3 | GO:0010138 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 1.4 | 5.6 | GO:0006226 | dUMP biosynthetic process(GO:0006226) pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 1.4 | 5.5 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 1.3 | 4.0 | GO:1990428 | miRNA transport(GO:1990428) |
| 1.3 | 27.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.2 | 3.6 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 1.2 | 3.6 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 1.2 | 4.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.1 | 3.4 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 1.1 | 7.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.0 | 6.2 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) positive regulation of protein localization to centrosome(GO:1904781) |
| 1.0 | 6.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.0 | 8.2 | GO:0006868 | glutamine transport(GO:0006868) |
| 1.0 | 7.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.0 | 4.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.9 | 2.7 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.9 | 2.7 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.9 | 6.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.9 | 8.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.9 | 3.5 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.9 | 9.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.8 | 2.5 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.8 | 4.0 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.8 | 6.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.8 | 2.4 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.8 | 5.3 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.8 | 4.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.8 | 7.6 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.8 | 3.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.7 | 3.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.7 | 16.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.7 | 2.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.7 | 2.8 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.7 | 9.0 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.7 | 15.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.7 | 5.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.6 | 3.8 | GO:1903336 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.6 | 5.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.6 | 2.5 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.6 | 6.9 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.6 | 2.5 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.6 | 5.5 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.6 | 4.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.6 | 4.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.6 | 4.2 | GO:0071877 | negative regulation of phospholipase activity(GO:0010519) regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.6 | 2.4 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.6 | 19.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.6 | 1.7 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.6 | 4.0 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.6 | 1.1 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.6 | 2.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.6 | 4.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.5 | 1.6 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.5 | 2.7 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.5 | 2.7 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.5 | 4.8 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.5 | 2.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 1.5 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.5 | 31.7 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.5 | 0.5 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.5 | 5.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.5 | 1.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 5.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.5 | 7.6 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.5 | 2.3 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.5 | 1.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.4 | 4.0 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.4 | 3.5 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.4 | 1.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.4 | 1.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.4 | 2.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 1.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.4 | 1.6 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 4.1 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.4 | 2.0 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.4 | 1.6 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.4 | 4.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.4 | 7.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.4 | 1.2 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.4 | 4.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.4 | 1.9 | GO:0060356 | leucine import(GO:0060356) |
| 0.4 | 3.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.4 | 5.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.4 | 1.1 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.4 | 1.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.4 | 4.1 | GO:0085020 | negative regulation of protein export from nucleus(GO:0046826) protein K6-linked ubiquitination(GO:0085020) |
| 0.4 | 3.0 | GO:0032202 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.4 | 1.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.4 | 1.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.4 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 2.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 2.7 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.3 | 1.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 3.4 | GO:1901374 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.3 | 6.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 1.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 1.2 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.3 | 5.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 1.8 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 2.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 0.9 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.3 | 6.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.3 | 2.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.3 | 1.4 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.3 | 3.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 0.8 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.3 | 2.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 1.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.3 | 2.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.3 | 8.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 0.8 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.3 | 2.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.3 | 5.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 0.8 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 | 2.8 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.3 | 3.8 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.3 | 1.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 4.5 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 5.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 1.0 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.2 | 1.9 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 5.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.6 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 | 1.6 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 1.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 0.4 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.2 | 0.2 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.2 | 4.8 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 1.5 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 1.6 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 | 10.9 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 0.2 | GO:0002649 | regulation of tolerance induction to self antigen(GO:0002649) |
| 0.2 | 1.2 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.2 | 1.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 | 0.4 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.2 | 8.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 0.4 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.2 | 1.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 1.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.2 | 0.4 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.2 | 3.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 3.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 0.9 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.2 | 2.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 0.9 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.2 | 19.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 1.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 1.1 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 2.2 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.2 | 1.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.2 | 3.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 4.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 0.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 1.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.9 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 2.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 8.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 4.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 11.9 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 | 0.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.4 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 3.7 | GO:1903205 | cell death in response to hydrogen peroxide(GO:0036474) regulation of hydrogen peroxide-induced cell death(GO:1903205) |
| 0.1 | 3.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.5 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 1.3 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 2.2 | GO:0032366 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.1 | 1.1 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.5 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 4.6 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 1.0 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 2.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 3.8 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.8 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 15.0 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 2.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.7 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 2.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 2.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.8 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 10.7 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.1 | 0.3 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.2 | GO:0002652 | regulation of tolerance induction dependent upon immune response(GO:0002652) |
| 0.1 | 0.4 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 0.2 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 4.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.8 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 15.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 3.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.8 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.1 | 1.5 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 2.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 7.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.9 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 1.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 1.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.8 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 6.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.1 | 3.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.8 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 1.0 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 2.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 1.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 2.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 2.9 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.2 | GO:0043382 | defense response to nematode(GO:0002215) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 2.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 31.0 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 3.3 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0010925 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 1.4 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 1.0 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 2.0 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) mast cell degranulation(GO:0043303) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 1.9 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 17.6 | GO:0051726 | regulation of cell cycle(GO:0051726) |
| 0.0 | 3.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.9 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 1.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 3.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 1.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.1 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.0 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 2.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 2.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 2.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 28.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 2.3 | 39.4 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 2.3 | 6.9 | GO:0000811 | GINS complex(GO:0000811) |
| 1.9 | 5.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.4 | 9.9 | GO:0071914 | prominosome(GO:0071914) |
| 1.1 | 6.9 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 1.1 | 12.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 1.1 | 9.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 1.0 | 8.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.9 | 15.7 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.9 | 10.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.8 | 39.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.8 | 21.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.8 | 14.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.8 | 4.8 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.8 | 14.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.8 | 7.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.7 | 4.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.7 | 5.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.6 | 3.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.6 | 1.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.6 | 5.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.6 | 5.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 3.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.5 | 2.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.5 | 1.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.5 | 4.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.5 | 2.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.5 | 1.9 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.5 | 7.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.5 | 5.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.5 | 2.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 10.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 1.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.4 | 9.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 2.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 3.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.4 | 2.6 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.4 | 4.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.4 | 1.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 5.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 1.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 9.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 10.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 4.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 1.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 4.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 3.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 19.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 10.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 2.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 3.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 1.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 3.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 16.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.2 | 16.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 5.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 4.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 9.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 0.5 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 0.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 1.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 5.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 1.1 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 4.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 2.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 1.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 3.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 2.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 19.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 2.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 3.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 7.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 19.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.1 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 2.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 2.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 3.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.4 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.9 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.1 | 1.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 2.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 7.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 3.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 9.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 18.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 2.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.5 | GO:0005883 | neurofilament(GO:0005883) C-fiber(GO:0044299) |
| 0.1 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 11.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 1.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 5.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 4.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 2.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 7.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 2.8 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 2.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 3.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 5.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 8.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 4.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 4.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 28.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 5.4 | 21.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 2.0 | 8.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 1.8 | 5.4 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 1.7 | 5.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 1.6 | 4.8 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 1.6 | 4.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.4 | 7.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.4 | 5.5 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 1.3 | 16.0 | GO:0008061 | chitin binding(GO:0008061) |
| 1.3 | 7.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.3 | 7.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 1.2 | 10.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 1.2 | 11.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.2 | 17.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.2 | 8.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 1.1 | 4.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.0 | 4.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.0 | 6.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.0 | 10.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.0 | 4.8 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.9 | 8.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.9 | 5.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.9 | 6.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.9 | 2.7 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.9 | 8.8 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.8 | 4.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.8 | 3.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.8 | 3.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.8 | 39.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.7 | 4.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.7 | 2.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.7 | 2.8 | GO:0038100 | nodal binding(GO:0038100) |
| 0.7 | 10.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.7 | 5.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.7 | 4.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.6 | 1.9 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.6 | 6.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.6 | 4.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.6 | 7.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.6 | 5.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 3.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.5 | 2.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 1.6 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.5 | 7.2 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.5 | 5.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.5 | 11.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 1.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.5 | 2.4 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.5 | 3.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.4 | 1.3 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 6.9 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.4 | 1.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.4 | 6.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 3.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.4 | 3.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 2.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 1.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.4 | 2.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 2.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 2.8 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 1.7 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 2.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 1.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 6.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 9.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.3 | 2.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 1.5 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.3 | 1.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 1.4 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.3 | 6.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 3.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.3 | 1.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 2.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 0.8 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.3 | 1.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 7.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 2.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 3.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 1.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 14.4 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 14.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 1.7 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 0.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 3.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 4.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 1.3 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.2 | 5.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 3.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 4.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.5 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.2 | 0.6 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 0.8 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.2 | 2.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 3.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 1.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 5.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 11.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 0.6 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 3.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 4.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 5.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 1.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 2.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 1.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 0.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 6.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 1.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 1.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 2.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 29.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 5.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 9.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 8.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 7.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 8.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 10.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.8 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 14.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 1.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 3.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 5.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 6.9 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 1.0 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 4.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 3.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.9 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 8.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 5.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 10.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 7.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 7.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 2.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 4.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 12.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 4.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 20.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 2.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 3.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.4 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 1.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 3.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 3.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 15.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.6 | 38.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.6 | 10.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 10.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 19.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 10.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 7.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 16.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 10.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 11.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 6.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 7.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 10.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 11.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 15.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 4.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 17.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 4.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 6.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 5.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 10.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 4.2 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 2.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 13.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 3.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 27.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 14.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.6 | 7.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 10.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.5 | 10.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.5 | 19.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.5 | 10.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 15.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 19.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 6.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 7.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 34.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 19.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.3 | 4.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 6.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 15.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.3 | 3.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 8.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 9.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 5.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 6.6 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.3 | 3.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 7.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 7.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 31.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 2.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 14.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 4.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 1.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 1.6 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 4.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 5.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 3.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 1.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 3.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.1 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 6.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 3.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 4.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 6.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 1.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |