Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Neurod2_Bhlha15_Bhlhe22_Olig1
Z-value: 1.21
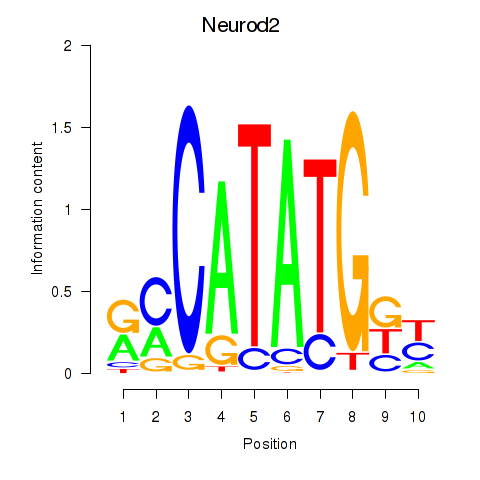
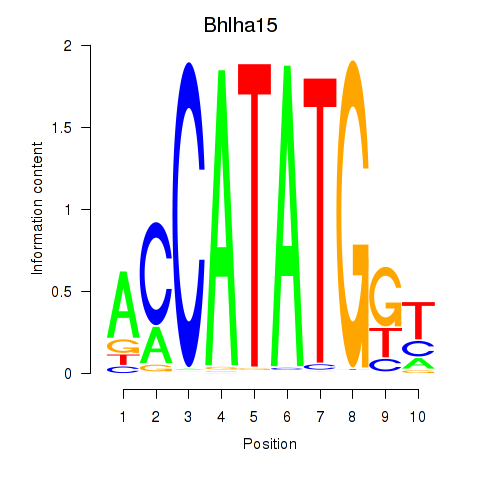
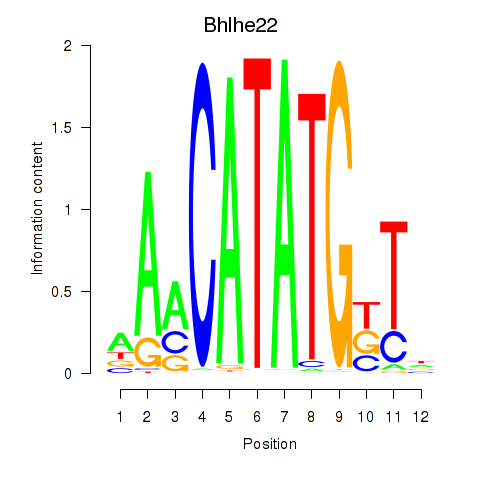
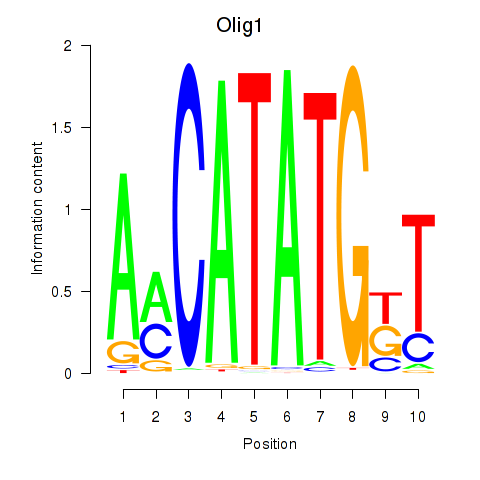
Transcription factors associated with Neurod2_Bhlha15_Bhlhe22_Olig1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurod2
|
ENSMUSG00000038255.7 | neurogenic differentiation 2 |
|
Bhlha15
|
ENSMUSG00000052271.8 | basic helix-loop-helix family, member a15 |
|
Bhlhe22
|
ENSMUSG00000025128.8 | basic helix-loop-helix family, member e22 |
|
Olig1
|
ENSMUSG00000046160.7 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Olig1 | mm39_v1_chr16_+_91066602_91066667 | 0.72 | 8.4e-07 | Click! |
| Bhlha15 | mm39_v1_chr5_+_144127102_144127152 | -0.52 | 1.2e-03 | Click! |
| Bhlhe22 | mm39_v1_chr3_+_18108313_18108338 | -0.43 | 9.2e-03 | Click! |
| Neurod2 | mm39_v1_chr11_-_98220466_98220482 | -0.21 | 2.1e-01 | Click! |
Activity profile of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
Sorted Z-values of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_87697155 | 21.77 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr8_+_86219191 | 10.10 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr15_-_75963446 | 7.48 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr6_+_124489364 | 6.96 |
ENSMUST00000068593.9
|
C1ra
|
complement component 1, r subcomponent A |
| chr6_+_90439596 | 5.65 |
ENSMUST00000203039.3
|
Klf15
|
Kruppel-like factor 15 |
| chr6_+_90439544 | 5.61 |
ENSMUST00000032174.12
|
Klf15
|
Kruppel-like factor 15 |
| chr9_+_72892850 | 5.28 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr9_-_72892617 | 4.97 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr10_+_127637015 | 4.54 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr8_+_95564949 | 4.53 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr15_-_76010736 | 4.48 |
ENSMUST00000054022.12
ENSMUST00000089654.4 |
BC024139
|
cDNA sequence BC024139 |
| chr10_+_127702326 | 4.28 |
ENSMUST00000092058.4
|
Rdh16f2
|
RDH16 family member 2 |
| chr1_-_150268470 | 3.97 |
ENSMUST00000006167.13
ENSMUST00000097547.10 |
Odr4
|
odr4 GPCR localization factor homolog |
| chr11_-_69696428 | 3.75 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr2_+_155224105 | 3.70 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr18_+_12776358 | 3.61 |
ENSMUST00000234966.2
ENSMUST00000025294.9 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr9_-_110571645 | 3.54 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr1_-_139487951 | 3.26 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr3_+_146302832 | 3.05 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr10_+_127595639 | 2.78 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr10_-_127724557 | 2.78 |
ENSMUST00000047199.5
|
Rdh7
|
retinol dehydrogenase 7 |
| chr4_+_85972125 | 2.72 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr5_+_137979763 | 2.65 |
ENSMUST00000035390.7
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr19_-_46661321 | 2.54 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr8_+_26091607 | 2.48 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr19_+_26725589 | 2.46 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_104125226 | 2.46 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr19_-_46661501 | 2.40 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr5_-_104125192 | 2.40 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr5_-_104125270 | 2.35 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_+_167445815 | 2.32 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr1_+_165591315 | 1.99 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr9_+_72892786 | 1.90 |
ENSMUST00000156879.8
|
Ccpg1
|
cell cycle progression 1 |
| chrX_+_165127688 | 1.90 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr8_-_106660470 | 1.89 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr9_+_66853343 | 1.83 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr1_+_177272215 | 1.78 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr16_-_45830575 | 1.75 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_+_67665434 | 1.72 |
ENSMUST00000181566.2
|
Gsg1l2
|
GSG1-like 2 |
| chr8_-_5155347 | 1.69 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr10_-_126877382 | 1.69 |
ENSMUST00000116231.4
|
Eef1akmt3
|
EEF1A lysine methyltransferase 3 |
| chr2_+_69500444 | 1.63 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr9_-_121745354 | 1.56 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr11_+_43046476 | 1.55 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr3_-_146302343 | 1.55 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr9_-_99599312 | 1.53 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr4_+_150938376 | 1.51 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr15_+_99870714 | 1.51 |
ENSMUST00000230956.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr8_+_26092252 | 1.42 |
ENSMUST00000136107.9
ENSMUST00000146919.8 ENSMUST00000139966.8 ENSMUST00000142395.8 ENSMUST00000143445.2 |
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr4_+_80828883 | 1.42 |
ENSMUST00000055922.4
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr2_+_71884943 | 1.42 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chrY_+_2900989 | 1.40 |
ENSMUST00000187842.7
|
Gm10352
|
predicted gene 10352 |
| chr16_+_43056218 | 1.40 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chrY_+_3771673 | 1.39 |
ENSMUST00000186140.7
|
Gm3376
|
predicted gene 3376 |
| chrY_-_2796205 | 1.39 |
ENSMUST00000187482.2
|
Gm4064
|
predicted gene 4064 |
| chrY_-_3306449 | 1.39 |
ENSMUST00000189592.7
|
Gm21677
|
predicted gene, 21677 |
| chrY_-_3378783 | 1.39 |
ENSMUST00000187277.7
|
Gm21704
|
predicted gene, 21704 |
| chr17_-_37300396 | 1.39 |
ENSMUST00000169189.8
|
H2-M5
|
histocompatibility 2, M region locus 5 |
| chrY_+_2862139 | 1.39 |
ENSMUST00000189964.7
ENSMUST00000188114.2 |
Gm10256
|
predicted gene 10256 |
| chrY_+_2830680 | 1.39 |
ENSMUST00000171534.8
ENSMUST00000100360.5 ENSMUST00000179404.8 |
Rbmy
Gm10256
|
RNA binding motif protein, Y chromosome predicted gene 10256 |
| chr5_-_66776095 | 1.38 |
ENSMUST00000162366.8
ENSMUST00000162994.8 ENSMUST00000159512.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr2_+_118603247 | 1.36 |
ENSMUST00000061360.4
ENSMUST00000130293.8 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr16_+_16031182 | 1.27 |
ENSMUST00000039408.3
|
Pkp2
|
plakophilin 2 |
| chrY_-_3410148 | 1.24 |
ENSMUST00000190283.7
ENSMUST00000188091.7 ENSMUST00000169382.3 |
Gm21708
Gm21704
|
predicted gene, 21708 predicted gene, 21704 |
| chrX_-_74423647 | 1.23 |
ENSMUST00000114085.9
|
F8
|
coagulation factor VIII |
| chr4_+_11579648 | 1.22 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr14_+_53797089 | 1.20 |
ENSMUST00000200101.2
ENSMUST00000103653.3 |
Trav15-1-dv6-1
|
T cell receptor alpha variable 15-1-DV6-1 |
| chr9_-_58447997 | 1.20 |
ENSMUST00000216629.2
|
Cd276
|
CD276 antigen |
| chr9_+_72892693 | 1.16 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr10_-_127358231 | 1.15 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr11_+_32592707 | 1.12 |
ENSMUST00000109366.8
ENSMUST00000093205.13 ENSMUST00000076383.8 |
Fbxw11
|
F-box and WD-40 domain protein 11 |
| chr13_+_8935974 | 1.12 |
ENSMUST00000177397.8
ENSMUST00000177400.8 ENSMUST00000177447.2 |
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr10_-_127358300 | 1.11 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr10_+_127595590 | 1.10 |
ENSMUST00000073639.6
|
Rdh1
|
retinol dehydrogenase 1 (all trans) |
| chr5_-_66775979 | 1.08 |
ENSMUST00000162382.8
ENSMUST00000159786.8 ENSMUST00000162349.8 ENSMUST00000087256.12 ENSMUST00000160103.8 ENSMUST00000160870.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr14_-_10185523 | 1.07 |
ENSMUST00000223702.2
ENSMUST00000223762.2 ENSMUST00000112669.10 ENSMUST00000163392.3 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chr2_+_127429125 | 1.07 |
ENSMUST00000028852.13
|
Mrps5
|
mitochondrial ribosomal protein S5 |
| chr4_-_108158242 | 1.06 |
ENSMUST00000043616.7
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chr2_+_59442378 | 1.06 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr7_+_130633776 | 1.06 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr1_+_15357478 | 1.05 |
ENSMUST00000175681.3
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr14_-_10185481 | 1.05 |
ENSMUST00000225640.2
ENSMUST00000225294.2 ENSMUST00000224389.2 ENSMUST00000223927.2 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chr10_+_127256192 | 1.05 |
ENSMUST00000171434.8
|
R3hdm2
|
R3H domain containing 2 |
| chr6_-_116050081 | 1.04 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr15_+_99870787 | 1.02 |
ENSMUST00000231160.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr6_-_148732893 | 1.01 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr13_+_8935537 | 1.00 |
ENSMUST00000169314.9
|
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr2_-_101479846 | 0.98 |
ENSMUST00000078494.6
ENSMUST00000160722.8 ENSMUST00000160037.8 |
Rag1
Iftap
|
recombination activating 1 intraflagellar transport associated protein |
| chr1_+_93062962 | 0.96 |
ENSMUST00000027491.7
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr1_+_177272297 | 0.96 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr11_+_70735751 | 0.95 |
ENSMUST00000177731.8
ENSMUST00000108533.10 ENSMUST00000081362.13 ENSMUST00000178245.2 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr13_-_101831020 | 0.90 |
ENSMUST00000185795.2
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr14_+_67470735 | 0.88 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr17_+_3532455 | 0.87 |
ENSMUST00000227604.2
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr9_-_48391838 | 0.87 |
ENSMUST00000216470.2
ENSMUST00000217037.2 ENSMUST00000034524.5 ENSMUST00000213895.2 |
Rexo2
|
RNA exonuclease 2 |
| chr6_+_51521493 | 0.86 |
ENSMUST00000179365.8
ENSMUST00000114439.8 |
Snx10
|
sorting nexin 10 |
| chr9_+_37313193 | 0.85 |
ENSMUST00000214185.3
|
Robo4
|
roundabout guidance receptor 4 |
| chr14_-_66191177 | 0.85 |
ENSMUST00000042046.5
|
Scara3
|
scavenger receptor class A, member 3 |
| chr15_+_101013704 | 0.84 |
ENSMUST00000229954.2
|
Ankrd33
|
ankyrin repeat domain 33 |
| chr2_-_29142965 | 0.83 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr11_-_96720738 | 0.82 |
ENSMUST00000107657.8
ENSMUST00000081775.12 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr5_-_135991117 | 0.81 |
ENSMUST00000111150.2
ENSMUST00000054895.4 |
Ssc4d
|
scavenger receptor cysteine rich family, 4 domains |
| chr10_+_110756031 | 0.81 |
ENSMUST00000220409.2
ENSMUST00000219502.2 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr14_+_67470884 | 0.81 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr14_+_74969737 | 0.77 |
ENSMUST00000022573.17
ENSMUST00000175712.8 ENSMUST00000176957.8 |
Esd
|
esterase D/formylglutathione hydrolase |
| chr5_-_5529119 | 0.76 |
ENSMUST00000115447.2
|
Pttg1ip2
|
PTTG1IP family member 2 |
| chr19_-_4384029 | 0.76 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr6_-_122317156 | 0.75 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr11_-_75686874 | 0.75 |
ENSMUST00000021209.8
|
Doc2b
|
double C2, beta |
| chr17_+_35345292 | 0.74 |
ENSMUST00000061859.7
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr2_+_111491764 | 0.72 |
ENSMUST00000213511.2
ENSMUST00000207228.3 ENSMUST00000208983.2 |
Olfr1299
Gm44840
|
olfactory receptor 1299 predicted gene 44840 |
| chr9_+_89093210 | 0.71 |
ENSMUST00000118870.8
ENSMUST00000085256.8 |
Mthfs
|
5, 10-methenyltetrahydrofolate synthetase |
| chr2_+_65499097 | 0.68 |
ENSMUST00000200829.4
|
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr13_-_67480588 | 0.67 |
ENSMUST00000109735.9
ENSMUST00000168892.9 |
Zfp595
|
zinc finger protein 595 |
| chrX_+_140258381 | 0.66 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr9_+_24194729 | 0.66 |
ENSMUST00000154644.2
|
Npsr1
|
neuropeptide S receptor 1 |
| chr3_-_84167119 | 0.63 |
ENSMUST00000107691.8
|
Trim2
|
tripartite motif-containing 2 |
| chr17_-_46798566 | 0.63 |
ENSMUST00000047034.9
|
Ttbk1
|
tau tubulin kinase 1 |
| chr6_+_91881193 | 0.61 |
ENSMUST00000205686.2
|
4930590J08Rik
|
RIKEN cDNA 4930590J08 gene |
| chr11_-_98220466 | 0.61 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr13_+_81034214 | 0.59 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr19_+_5100475 | 0.58 |
ENSMUST00000225427.2
|
Rin1
|
Ras and Rab interactor 1 |
| chr7_-_103420801 | 0.58 |
ENSMUST00000106878.3
|
Olfr69
|
olfactory receptor 69 |
| chr16_-_33916354 | 0.57 |
ENSMUST00000114973.9
ENSMUST00000232157.2 ENSMUST00000114964.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr11_-_53313950 | 0.57 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr18_-_35841435 | 0.57 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr12_+_10419967 | 0.55 |
ENSMUST00000143739.9
ENSMUST00000002456.10 ENSMUST00000219826.2 ENSMUST00000217944.2 ENSMUST00000218339.3 ENSMUST00000118657.8 ENSMUST00000223534.2 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr9_-_88601866 | 0.55 |
ENSMUST00000113110.5
|
Mthfsl
|
5, 10-methenyltetrahydrofolate synthetase-like |
| chr13_-_67480562 | 0.54 |
ENSMUST00000171466.2
|
Zfp595
|
zinc finger protein 595 |
| chr7_+_46636562 | 0.54 |
ENSMUST00000185832.2
|
Gm9999
|
predicted gene 9999 |
| chr2_+_91541245 | 0.53 |
ENSMUST00000142692.2
ENSMUST00000090608.6 |
Harbi1
|
harbinger transposase derived 1 |
| chr2_+_61876956 | 0.53 |
ENSMUST00000112480.3
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_-_121637505 | 0.52 |
ENSMUST00000138157.8
|
Frmd5
|
FERM domain containing 5 |
| chr3_+_63148887 | 0.52 |
ENSMUST00000194324.6
|
Mme
|
membrane metallo endopeptidase |
| chr14_+_30376310 | 0.51 |
ENSMUST00000064230.16
|
Rft1
|
RFT1 homolog |
| chr14_-_10185787 | 0.51 |
ENSMUST00000225871.2
|
Gm49355
|
predicted gene, 49355 |
| chr3_+_105821450 | 0.50 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr16_-_44153288 | 0.49 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr3_-_107240989 | 0.49 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr1_-_80439165 | 0.49 |
ENSMUST00000211023.2
|
Gm45261
|
predicted gene 45261 |
| chr5_-_25047577 | 0.49 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr8_-_107064615 | 0.49 |
ENSMUST00000067512.8
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr2_+_91541197 | 0.49 |
ENSMUST00000128140.2
ENSMUST00000140183.2 |
Harbi1
|
harbinger transposase derived 1 |
| chr3_-_92481033 | 0.48 |
ENSMUST00000053107.6
|
Ivl
|
involucrin |
| chr6_-_128414616 | 0.48 |
ENSMUST00000151796.3
|
Fkbp4
|
FK506 binding protein 4 |
| chr10_+_59715439 | 0.47 |
ENSMUST00000142819.8
ENSMUST00000020309.7 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr8_-_34614187 | 0.47 |
ENSMUST00000033910.9
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr15_+_30172716 | 0.45 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr2_-_26962187 | 0.45 |
ENSMUST00000009358.9
|
Mymk
|
myomaker, myoblast fusion factor |
| chr8_+_72863330 | 0.45 |
ENSMUST00000209675.2
|
Olfr374
|
olfactory receptor 374 |
| chr16_-_44153498 | 0.43 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr11_+_115494751 | 0.43 |
ENSMUST00000058109.9
|
Mrps7
|
mitchondrial ribosomal protein S7 |
| chr1_+_157334347 | 0.43 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr17_-_18075450 | 0.43 |
ENSMUST00000003762.8
|
Has1
|
hyaluronan synthase 1 |
| chr11_+_117123107 | 0.43 |
ENSMUST00000106354.9
|
Septin9
|
septin 9 |
| chr4_-_49681954 | 0.42 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chr1_+_157334298 | 0.42 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr5_+_123648523 | 0.42 |
ENSMUST00000031384.6
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr7_+_30157704 | 0.41 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr7_-_133203838 | 0.41 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr9_-_119654522 | 0.40 |
ENSMUST00000070617.8
|
Scn11a
|
sodium channel, voltage-gated, type XI, alpha |
| chr2_-_130480014 | 0.40 |
ENSMUST00000089561.10
ENSMUST00000110260.8 |
Lzts3
|
leucine zipper, putative tumor suppressor family member 3 |
| chr10_+_59715378 | 0.39 |
ENSMUST00000147914.8
ENSMUST00000146590.8 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chrX_-_140508177 | 0.38 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr10_-_117582259 | 0.37 |
ENSMUST00000079041.7
|
Slc35e3
|
solute carrier family 35, member E3 |
| chr13_+_67979225 | 0.37 |
ENSMUST00000221266.2
|
Gm10037
|
predicted gene 10037 |
| chr7_-_126062272 | 0.35 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr10_-_95337783 | 0.35 |
ENSMUST00000075829.3
ENSMUST00000217777.2 ENSMUST00000218893.2 |
Mrpl42
|
mitochondrial ribosomal protein L42 |
| chr1_+_177273226 | 0.35 |
ENSMUST00000077225.8
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_+_10056631 | 0.35 |
ENSMUST00000207792.3
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr2_+_61876923 | 0.35 |
ENSMUST00000054484.15
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr16_-_57113207 | 0.34 |
ENSMUST00000023434.15
ENSMUST00000120112.2 ENSMUST00000119407.8 |
Tmem30c
|
transmembrane protein 30C |
| chr15_+_98065039 | 0.34 |
ENSMUST00000031914.6
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr16_-_31133622 | 0.34 |
ENSMUST00000115230.2
ENSMUST00000130560.8 |
Apod
|
apolipoprotein D |
| chr2_+_86655007 | 0.34 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr16_-_19014062 | 0.33 |
ENSMUST00000197046.2
|
Iglc4
|
immunoglobulin lambda constant 4 |
| chr14_+_63235512 | 0.33 |
ENSMUST00000100492.5
|
Defb47
|
defensin beta 47 |
| chr11_-_6588637 | 0.33 |
ENSMUST00000102910.4
|
Wap
|
whey acidic protein |
| chr5_-_142594549 | 0.33 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr1_-_131441962 | 0.31 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr5_+_124621521 | 0.31 |
ENSMUST00000111453.2
|
Snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr18_+_38809771 | 0.30 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr18_+_62457275 | 0.30 |
ENSMUST00000027560.8
|
Htr4
|
5 hydroxytryptamine (serotonin) receptor 4 |
| chr2_+_65760477 | 0.29 |
ENSMUST00000176109.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr13_-_67523832 | 0.29 |
ENSMUST00000225787.2
ENSMUST00000172266.8 ENSMUST00000057070.9 |
Zfp456
|
zinc finger protein 456 |
| chr14_+_53550361 | 0.29 |
ENSMUST00000179997.2
|
Trav5n-4
|
T cell receptor alpha variable 5N-4 |
| chr19_+_45991907 | 0.29 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chr17_-_80884839 | 0.29 |
ENSMUST00000234349.2
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr6_+_97906760 | 0.28 |
ENSMUST00000101123.10
|
Mitf
|
melanogenesis associated transcription factor |
| chr5_-_90487583 | 0.28 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr17_+_71511642 | 0.28 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr10_-_25076008 | 0.28 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr2_+_154042291 | 0.27 |
ENSMUST00000028987.7
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr15_-_88863210 | 0.27 |
ENSMUST00000042594.13
ENSMUST00000109368.2 |
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) |
| chr17_+_38231439 | 0.27 |
ENSMUST00000216440.2
|
Olfr128
|
olfactory receptor 128 |
| chr9_+_37313287 | 0.27 |
ENSMUST00000115048.10
ENSMUST00000115046.9 ENSMUST00000102895.7 ENSMUST00000239486.2 |
Robo4
|
roundabout guidance receptor 4 |
| chr8_+_31579633 | 0.27 |
ENSMUST00000170204.8
|
Dusp26
|
dual specificity phosphatase 26 (putative) |
| chr17_+_37977879 | 0.26 |
ENSMUST00000215811.2
|
Olfr118
|
olfactory receptor 118 |
| chr13_-_99584091 | 0.26 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
| chr3_+_137770813 | 0.26 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurod2_Bhlha15_Bhlhe22_Olig1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 21.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 2.8 | 11.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 1.0 | 8.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.8 | 2.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 4.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 3.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.4 | 11.3 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.3 | 0.3 | GO:2000405 | negative regulation of T cell migration(GO:2000405) |
| 0.3 | 2.5 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 1.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 4.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.3 | 0.8 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.3 | 1.5 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.2 | 1.9 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 2.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 3.7 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 1.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.2 | 1.3 | GO:0002159 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) |
| 0.2 | 3.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 0.6 | GO:0090327 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 1.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.2 | 4.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 1.2 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.2 | 0.7 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.2 | 3.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.5 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.1 | 0.4 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 2.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 3.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 1.6 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 0.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.6 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.8 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.8 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.3 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.1 | 0.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.2 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 2.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.2 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.1 | 0.6 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.2 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.1 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 1.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 1.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.7 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 1.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 7.2 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.1 | 2.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 6.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.7 | GO:1900103 | negative regulation of heart rate(GO:0010459) positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0098923 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 3.5 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.4 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.9 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.8 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.1 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.5 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 1.9 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 3.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 1.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0006404 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.3 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.4 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 2.5 | GO:0001764 | neuron migration(GO:0001764) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 21.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 1.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 2.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 0.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.7 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 2.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 4.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 4.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 7.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.2 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.1 | 1.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 3.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 3.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 3.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 6.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 2.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 1.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.1 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.8 | 2.3 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.5 | 3.5 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.5 | 22.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 2.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.4 | 1.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.4 | 3.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 8.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 1.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.3 | 4.9 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 3.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 2.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 1.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.2 | 0.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.6 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.1 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 1.8 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 1.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 2.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 2.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 2.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 8.6 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.2 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 1.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 3.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 1.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.5 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 1.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 22.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 4.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 10.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 1.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |