Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
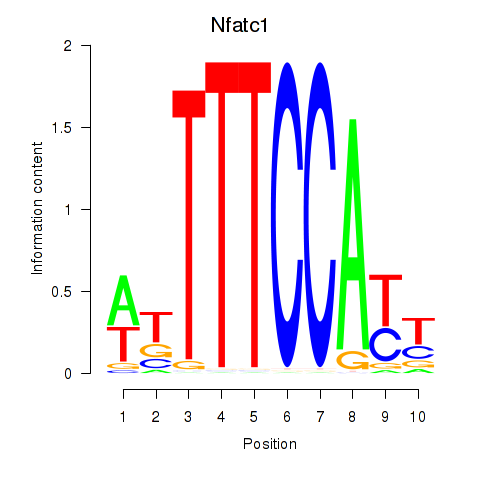
Results for Nfatc1
Z-value: 0.88
Transcription factors associated with Nfatc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc1
|
ENSMUSG00000033016.17 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | mm39_v1_chr18_-_80756273_80756295 | -0.03 | 8.4e-01 | Click! |
Activity profile of Nfatc1 motif
Sorted Z-values of Nfatc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41498716 | 4.70 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr7_-_142233270 | 4.37 |
ENSMUST00000162317.2
ENSMUST00000125933.2 ENSMUST00000105931.8 ENSMUST00000105930.8 ENSMUST00000105933.8 ENSMUST00000105932.2 ENSMUST00000000220.3 |
Ins2
|
insulin II |
| chr4_-_141553306 | 3.98 |
ENSMUST00000102481.4
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr19_+_58658779 | 3.57 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr19_+_52252735 | 3.00 |
ENSMUST00000039652.6
|
Ins1
|
insulin I |
| chr19_+_58658838 | 2.94 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr3_+_94284739 | 2.19 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr10_+_87696339 | 1.86 |
ENSMUST00000121161.8
|
Igf1
|
insulin-like growth factor 1 |
| chr3_+_94284812 | 1.85 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr1_-_180021039 | 1.72 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr1_-_180021218 | 1.70 |
ENSMUST00000159914.8
|
Coq8a
|
coenzyme Q8A |
| chr15_+_10314173 | 1.30 |
ENSMUST00000127467.3
|
Prlr
|
prolactin receptor |
| chr10_+_40505985 | 1.30 |
ENSMUST00000019977.8
ENSMUST00000214102.2 ENSMUST00000213503.2 ENSMUST00000213442.2 ENSMUST00000216830.2 |
Ddo
|
D-aspartate oxidase |
| chr18_-_39000056 | 1.30 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr9_-_51240201 | 1.25 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr10_+_87695886 | 1.16 |
ENSMUST00000062862.13
|
Igf1
|
insulin-like growth factor 1 |
| chr1_+_88139678 | 1.14 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr1_+_88022776 | 1.10 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr13_-_93774469 | 1.06 |
ENSMUST00000099309.6
|
Bhmt
|
betaine-homocysteine methyltransferase |
| chr1_-_66974492 | 0.99 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr7_-_126224848 | 0.97 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr18_+_45402018 | 0.94 |
ENSMUST00000183850.8
ENSMUST00000066890.14 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr2_-_110136074 | 0.89 |
ENSMUST00000046233.9
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr1_-_66974694 | 0.88 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr1_+_58152295 | 0.83 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr5_-_89605622 | 0.81 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr1_+_87998487 | 0.81 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chrX_-_74918709 | 0.80 |
ENSMUST00000114059.10
|
Pls3
|
plastin 3 (T-isoform) |
| chr15_-_3612703 | 0.79 |
ENSMUST00000069451.11
|
Ghr
|
growth hormone receptor |
| chr1_+_24717722 | 0.79 |
ENSMUST00000186096.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr6_-_21851827 | 0.79 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr5_+_90708962 | 0.77 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr11_+_98938137 | 0.77 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr1_+_24717711 | 0.76 |
ENSMUST00000191471.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr10_+_4561974 | 0.76 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr1_+_24717793 | 0.72 |
ENSMUST00000186190.2
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr15_+_25933632 | 0.71 |
ENSMUST00000228327.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr4_+_102446883 | 0.70 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr12_-_101784727 | 0.66 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr15_-_58828321 | 0.66 |
ENSMUST00000228067.2
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr2_+_67935015 | 0.64 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr14_+_55797468 | 0.62 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr10_-_53952686 | 0.61 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr15_-_89261242 | 0.60 |
ENSMUST00000023285.5
|
Tymp
|
thymidine phosphorylase |
| chr11_+_67090878 | 0.60 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr10_-_27492827 | 0.59 |
ENSMUST00000092639.12
|
Lama2
|
laminin, alpha 2 |
| chr5_-_44256562 | 0.59 |
ENSMUST00000165909.8
|
Prom1
|
prominin 1 |
| chr11_+_118324652 | 0.59 |
ENSMUST00000106286.3
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr14_+_55797443 | 0.58 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr5_-_44256528 | 0.58 |
ENSMUST00000196178.2
ENSMUST00000197750.5 |
Prom1
|
prominin 1 |
| chr11_-_99383938 | 0.56 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr17_-_32643067 | 0.56 |
ENSMUST00000237130.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr10_-_77002377 | 0.56 |
ENSMUST00000081654.13
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr14_+_5894220 | 0.56 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr15_-_76608009 | 0.53 |
ENSMUST00000036247.10
|
C030006K11Rik
|
RIKEN cDNA C030006K11 gene |
| chr16_-_45975440 | 0.51 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr19_+_43741550 | 0.51 |
ENSMUST00000153295.2
|
Cutc
|
cutC copper transporter |
| chr2_-_173117936 | 0.49 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr4_-_148021159 | 0.48 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr10_+_125802084 | 0.48 |
ENSMUST00000074807.8
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr8_+_46080746 | 0.47 |
ENSMUST00000145458.9
ENSMUST00000134321.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr17_-_48739874 | 0.47 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chrX_+_72527208 | 0.47 |
ENSMUST00000033741.15
ENSMUST00000169489.2 |
Bgn
|
biglycan |
| chr9_+_7692087 | 0.46 |
ENSMUST00000018767.8
|
Mmp7
|
matrix metallopeptidase 7 |
| chr1_-_66984178 | 0.45 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr14_-_4808744 | 0.45 |
ENSMUST00000022303.17
ENSMUST00000091471.12 |
Thrb
|
thyroid hormone receptor beta |
| chr18_+_31768126 | 0.44 |
ENSMUST00000234846.2
ENSMUST00000234772.2 |
Sap130
|
Sin3A associated protein |
| chr1_+_133965228 | 0.44 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr12_+_75355082 | 0.44 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr10_+_20024203 | 0.44 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr3_+_62327089 | 0.44 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr3_+_121761471 | 0.43 |
ENSMUST00000196479.5
ENSMUST00000197155.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr17_-_32643131 | 0.43 |
ENSMUST00000236386.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr7_+_27770655 | 0.42 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr18_+_64473091 | 0.42 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr10_+_21870565 | 0.42 |
ENSMUST00000020145.12
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr12_-_101785307 | 0.41 |
ENSMUST00000021603.9
|
Fbln5
|
fibulin 5 |
| chr3_-_148696155 | 0.41 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr19_+_43741431 | 0.40 |
ENSMUST00000026199.14
|
Cutc
|
cutC copper transporter |
| chr14_+_48683581 | 0.40 |
ENSMUST00000227440.2
ENSMUST00000124720.8 ENSMUST00000226422.2 ENSMUST00000226400.2 |
Tmem260
|
transmembrane protein 260 |
| chr1_+_4878460 | 0.40 |
ENSMUST00000131119.2
|
Lypla1
|
lysophospholipase 1 |
| chr11_+_114566257 | 0.39 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr9_-_58462720 | 0.39 |
ENSMUST00000165365.3
|
Cd276
|
CD276 antigen |
| chr19_+_43741513 | 0.39 |
ENSMUST00000112047.10
|
Cutc
|
cutC copper transporter |
| chr9_+_100525807 | 0.38 |
ENSMUST00000133388.2
|
Stag1
|
stromal antigen 1 |
| chr13_-_101831020 | 0.38 |
ENSMUST00000185795.2
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr1_-_65218217 | 0.38 |
ENSMUST00000097709.11
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr8_-_25528972 | 0.38 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr14_+_119025306 | 0.37 |
ENSMUST00000047761.13
ENSMUST00000071546.14 |
Cldn10
|
claudin 10 |
| chr8_+_46080840 | 0.37 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_+_91661074 | 0.36 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr19_-_10502468 | 0.36 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr17_-_25300112 | 0.36 |
ENSMUST00000024984.7
|
Tmem204
|
transmembrane protein 204 |
| chr14_-_33996185 | 0.35 |
ENSMUST00000227006.2
|
Shld2
|
shieldin complex subunit 2 |
| chr7_+_51530060 | 0.35 |
ENSMUST00000145049.2
|
Gas2
|
growth arrest specific 2 |
| chr5_-_103174794 | 0.33 |
ENSMUST00000128869.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr14_-_21102188 | 0.33 |
ENSMUST00000130370.3
ENSMUST00000224016.2 ENSMUST00000022371.10 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr16_-_28748410 | 0.33 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr1_-_189902868 | 0.33 |
ENSMUST00000177288.4
ENSMUST00000175916.8 |
Prox1
|
prospero homeobox 1 |
| chr18_+_36693024 | 0.32 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr7_+_51537645 | 0.32 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr3_+_137923521 | 0.32 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr5_-_88675700 | 0.32 |
ENSMUST00000087033.6
|
Jchain
|
immunoglobulin joining chain |
| chr9_-_58065800 | 0.31 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr11_+_93776965 | 0.31 |
ENSMUST00000063718.11
ENSMUST00000107854.9 |
Mbtd1
|
mbt domain containing 1 |
| chr17_+_3447465 | 0.31 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr6_-_124865155 | 0.30 |
ENSMUST00000024044.7
|
Cd4
|
CD4 antigen |
| chr4_-_44084167 | 0.30 |
ENSMUST00000030201.14
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr10_-_27492792 | 0.30 |
ENSMUST00000189575.2
|
Lama2
|
laminin, alpha 2 |
| chr10_-_93146937 | 0.30 |
ENSMUST00000008542.12
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr9_+_100525637 | 0.30 |
ENSMUST00000041418.13
|
Stag1
|
stromal antigen 1 |
| chr15_-_96917804 | 0.29 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr9_-_40366966 | 0.29 |
ENSMUST00000165104.8
ENSMUST00000045682.7 |
Gramd1b
|
GRAM domain containing 1B |
| chr14_+_52222283 | 0.29 |
ENSMUST00000093813.12
ENSMUST00000100639.11 ENSMUST00000182909.8 ENSMUST00000182760.8 ENSMUST00000182061.8 ENSMUST00000182193.2 |
Arhgef40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr8_+_46338557 | 0.29 |
ENSMUST00000210422.2
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr9_+_32027335 | 0.28 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr6_+_97187650 | 0.28 |
ENSMUST00000044681.7
|
Arl6ip5
|
ADP-ribosylation factor-like 6 interacting protein 5 |
| chr13_+_41013230 | 0.28 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr6_-_124888643 | 0.28 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr2_-_98497609 | 0.28 |
ENSMUST00000099683.2
|
Gm10800
|
predicted gene 10800 |
| chr5_-_105491795 | 0.28 |
ENSMUST00000171587.2
|
Gbp11
|
guanylate binding protein 11 |
| chr1_+_75336965 | 0.27 |
ENSMUST00000027409.10
|
Des
|
desmin |
| chr4_-_82423985 | 0.27 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr3_-_116505469 | 0.27 |
ENSMUST00000153108.6
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr6_-_88022172 | 0.27 |
ENSMUST00000203674.3
ENSMUST00000204126.2 ENSMUST00000113596.8 ENSMUST00000113600.10 |
Rab7
|
RAB7, member RAS oncogene family |
| chr3_-_104419128 | 0.27 |
ENSMUST00000199070.5
ENSMUST00000046316.11 ENSMUST00000198332.2 |
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr7_-_5128936 | 0.27 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr2_+_30127692 | 0.27 |
ENSMUST00000113654.8
ENSMUST00000095078.3 |
Lrrc8a
|
leucine rich repeat containing 8A VRAC subunit A |
| chr10_-_86843878 | 0.27 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr5_-_118382926 | 0.26 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr1_+_82210833 | 0.26 |
ENSMUST00000023262.6
|
Gm9747
|
predicted gene 9747 |
| chr8_+_46338498 | 0.26 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr10_-_12689345 | 0.26 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr3_-_57483330 | 0.26 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr19_+_10502612 | 0.26 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr5_+_17779273 | 0.26 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr17_+_6156738 | 0.26 |
ENSMUST00000142030.8
|
Tulp4
|
tubby like protein 4 |
| chr12_-_55539372 | 0.26 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr7_+_29794575 | 0.26 |
ENSMUST00000130526.2
ENSMUST00000108200.2 |
Zfp260
|
zinc finger protein 260 |
| chr4_-_141518202 | 0.25 |
ENSMUST00000038014.11
ENSMUST00000153880.2 |
Dnajc16
|
DnaJ heat shock protein family (Hsp40) member C16 |
| chr2_-_103133524 | 0.25 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr19_-_43741363 | 0.25 |
ENSMUST00000045562.6
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr1_-_135302971 | 0.25 |
ENSMUST00000041240.4
|
Shisa4
|
shisa family member 4 |
| chr19_+_10502679 | 0.25 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr16_+_31241085 | 0.25 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr1_-_155912216 | 0.25 |
ENSMUST00000027738.14
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr2_-_103133503 | 0.25 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr2_+_91090167 | 0.24 |
ENSMUST00000138470.2
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr17_+_15262510 | 0.24 |
ENSMUST00000226561.2
|
Ermard
|
ER membrane associated RNA degradation |
| chr13_-_23945189 | 0.24 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr9_+_105535585 | 0.24 |
ENSMUST00000186943.2
|
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr10_-_95251145 | 0.24 |
ENSMUST00000119917.2
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr14_+_67470735 | 0.24 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr9_+_89081407 | 0.24 |
ENSMUST00000138109.2
|
Gm29094
|
predicted gene 29094 |
| chr6_-_48422447 | 0.24 |
ENSMUST00000114564.8
|
Zfp467
|
zinc finger protein 467 |
| chr19_-_10502546 | 0.24 |
ENSMUST00000237827.2
|
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr4_-_94867300 | 0.23 |
ENSMUST00000075872.4
|
Mysm1
|
myb-like, SWIRM and MPN domains 1 |
| chrX_+_13147209 | 0.23 |
ENSMUST00000000804.7
|
Ddx3x
|
DEAD box helicase 3, X-linked |
| chr10_-_28862289 | 0.23 |
ENSMUST00000152363.8
ENSMUST00000015663.7 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr6_+_91661034 | 0.23 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chrX_+_165127688 | 0.23 |
ENSMUST00000112223.8
ENSMUST00000112224.8 ENSMUST00000112229.9 ENSMUST00000112228.8 ENSMUST00000112227.9 ENSMUST00000112226.3 |
Gpm6b
|
glycoprotein m6b |
| chr18_+_38383297 | 0.23 |
ENSMUST00000025314.7
ENSMUST00000236078.2 |
Dele1
|
DAP3 binding cell death enhancer 1 |
| chr8_+_70243813 | 0.22 |
ENSMUST00000034326.7
|
Atp13a1
|
ATPase type 13A1 |
| chr11_-_100151847 | 0.22 |
ENSMUST00000080893.7
|
Krt17
|
keratin 17 |
| chr15_-_79048674 | 0.22 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr9_-_50439942 | 0.22 |
ENSMUST00000214873.2
ENSMUST00000034570.7 ENSMUST00000213144.2 ENSMUST00000215416.2 |
Pts
|
6-pyruvoyl-tetrahydropterin synthase |
| chr9_+_3013140 | 0.22 |
ENSMUST00000143083.3
|
Gm10721
|
predicted gene 10721 |
| chr9_-_20432562 | 0.22 |
ENSMUST00000215908.2
ENSMUST00000068296.8 ENSMUST00000174462.8 ENSMUST00000213418.2 |
Zfp266
|
zinc finger protein 266 |
| chr4_-_42168603 | 0.22 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr11_-_106107132 | 0.22 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr10_-_95251327 | 0.21 |
ENSMUST00000172070.8
ENSMUST00000150432.8 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr4_-_42665763 | 0.21 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr18_+_65156620 | 0.21 |
ENSMUST00000080418.7
ENSMUST00000237854.2 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr9_+_100525501 | 0.20 |
ENSMUST00000146312.8
ENSMUST00000129269.8 |
Stag1
|
stromal antigen 1 |
| chr16_-_37360097 | 0.20 |
ENSMUST00000023525.9
|
Gtf2e1
|
general transcription factor II E, polypeptide 1 (alpha subunit) |
| chr13_+_83672654 | 0.20 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr6_+_88701810 | 0.20 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr5_+_120649233 | 0.20 |
ENSMUST00000068326.14
ENSMUST00000111890.9 ENSMUST00000076051.12 ENSMUST00000147496.2 |
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr11_-_106641454 | 0.20 |
ENSMUST00000068021.9
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr14_-_31503869 | 0.20 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr6_+_88701578 | 0.20 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr4_-_82423944 | 0.20 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr4_-_12087911 | 0.20 |
ENSMUST00000050686.10
|
Tmem67
|
transmembrane protein 67 |
| chr14_-_54843366 | 0.20 |
ENSMUST00000022786.6
|
4931414P19Rik
|
RIKEN cDNA 4931414P19 gene |
| chr1_-_91386976 | 0.20 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2 |
| chr14_+_25842597 | 0.19 |
ENSMUST00000112364.8
|
Anxa11
|
annexin A11 |
| chr1_-_164763091 | 0.19 |
ENSMUST00000027860.8
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr11_+_93776650 | 0.19 |
ENSMUST00000107853.8
ENSMUST00000107850.8 |
Mbtd1
|
mbt domain containing 1 |
| chr4_+_86666764 | 0.19 |
ENSMUST00000045512.15
ENSMUST00000082026.14 |
Dennd4c
|
DENN/MADD domain containing 4C |
| chr1_+_180158035 | 0.19 |
ENSMUST00000070181.7
|
Itpkb
|
inositol 1,4,5-trisphosphate 3-kinase B |
| chr7_+_30252687 | 0.19 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr19_-_34231600 | 0.19 |
ENSMUST00000238147.2
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr2_+_18681812 | 0.19 |
ENSMUST00000028071.13
|
Bmi1
|
Bmi1 polycomb ring finger oncogene |
| chr14_+_67470884 | 0.19 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chrX_+_142447361 | 0.19 |
ENSMUST00000126592.8
ENSMUST00000156449.8 ENSMUST00000155215.8 ENSMUST00000112865.8 |
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr3_-_88679881 | 0.19 |
ENSMUST00000090945.5
|
Syt11
|
synaptotagmin XI |
| chr18_+_37813286 | 0.19 |
ENSMUST00000192931.2
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chr5_+_104350475 | 0.18 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr13_+_67276253 | 0.18 |
ENSMUST00000052716.8
|
Zfp759
|
zinc finger protein 759 |
| chr14_+_25842565 | 0.18 |
ENSMUST00000022416.15
|
Anxa11
|
annexin A11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.6 | 2.3 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.4 | 1.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.4 | 3.0 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.4 | 1.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.4 | 4.0 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.4 | 1.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.3 | 6.5 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 | 1.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.2 | 0.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 5.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 0.6 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.2 | 0.2 | GO:2000554 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.2 | 1.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.2 | 1.0 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.2 | 0.8 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.2 | 0.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.6 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 3.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 3.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.9 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.6 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.2 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.9 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.3 | GO:0061295 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.7 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.9 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.3 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.1 | 0.2 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.1 | 0.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.7 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.4 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.6 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.2 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.4 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.4 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.2 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 1.0 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 1.0 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.8 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.3 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 1.0 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0043382 | regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:1904733 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.8 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0035874 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.2 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0071638 | negative regulation of T-helper 2 cell differentiation(GO:0045629) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.6 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.0 | 0.4 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:1901561 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.8 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.3 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.3 | GO:0051044 | regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 2.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 2.1 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.4 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) chaperone-mediated protein transport(GO:0072321) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 0.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 4.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 1.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.9 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 6.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 1.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.3 | 0.8 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.3 | 1.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 0.8 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 1.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 2.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.8 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.2 | 0.6 | GO:0001761 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.2 | 8.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 1.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.6 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.6 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 3.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 0.2 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 3.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.0 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 9.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0052597 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.9 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.0 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 8.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 7.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 4.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |