Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
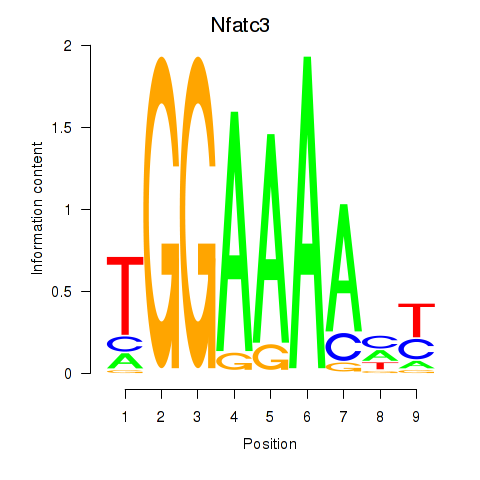
Results for Nfatc3
Z-value: 1.38
Transcription factors associated with Nfatc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc3
|
ENSMUSG00000031902.11 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc3 | mm39_v1_chr8_+_106786190_106786265 | -0.26 | 1.2e-01 | Click! |
Activity profile of Nfatc3 motif
Sorted Z-values of Nfatc3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_58658779 | 16.55 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr19_+_58658838 | 14.32 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr15_+_9335636 | 10.91 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr1_-_180021218 | 10.10 |
ENSMUST00000159914.8
|
Coq8a
|
coenzyme Q8A |
| chr15_+_76579885 | 9.87 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr4_-_107164315 | 9.60 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr15_+_10249646 | 8.61 |
ENSMUST00000134410.8
|
Prlr
|
prolactin receptor |
| chr9_-_48516447 | 8.19 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr19_-_7943365 | 7.90 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr14_+_40827108 | 7.14 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr15_+_10224052 | 7.05 |
ENSMUST00000128450.8
ENSMUST00000148257.8 ENSMUST00000128921.8 |
Prlr
|
prolactin receptor |
| chr1_-_180021039 | 6.90 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr14_+_40827317 | 6.48 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr1_-_150341911 | 6.16 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr1_+_88139678 | 6.16 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr10_+_87696339 | 5.65 |
ENSMUST00000121161.8
|
Igf1
|
insulin-like growth factor 1 |
| chr4_-_62005498 | 5.14 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr12_+_8027767 | 5.06 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr11_+_72326337 | 4.97 |
ENSMUST00000076443.10
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr1_+_88015524 | 4.81 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr10_+_21253190 | 4.80 |
ENSMUST00000042699.14
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr7_-_99276310 | 4.71 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr1_+_87998487 | 4.53 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr18_+_87774402 | 4.41 |
ENSMUST00000091776.7
|
Gm5096
|
predicted gene 5096 |
| chr14_+_40826970 | 4.32 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr3_+_94284739 | 4.21 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr6_-_21851827 | 4.16 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr15_-_96917804 | 4.07 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr7_+_26508305 | 3.99 |
ENSMUST00000040944.9
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr3_+_94284812 | 3.77 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr1_+_88022776 | 3.75 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr11_+_72326358 | 3.70 |
ENSMUST00000108499.2
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr19_-_8382424 | 3.60 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr10_+_4561974 | 3.53 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr1_+_58152295 | 3.25 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr5_-_89605622 | 3.22 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr14_+_55797443 | 3.10 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr13_+_30520416 | 3.09 |
ENSMUST00000222503.2
ENSMUST00000222370.2 ENSMUST00000066412.8 ENSMUST00000223201.2 |
Agtr1a
|
angiotensin II receptor, type 1a |
| chr14_+_55797468 | 3.08 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr11_+_98938137 | 3.02 |
ENSMUST00000140772.2
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr3_+_137923521 | 2.93 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_+_8027640 | 2.88 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr18_-_35760260 | 2.82 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr17_+_24955613 | 2.82 |
ENSMUST00000115262.9
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr6_+_91661074 | 2.81 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr18_-_60866186 | 2.77 |
ENSMUST00000237885.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr15_+_3300249 | 2.70 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr3_+_121761471 | 2.70 |
ENSMUST00000196479.5
ENSMUST00000197155.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr17_+_24955647 | 2.69 |
ENSMUST00000101800.7
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr12_+_37291728 | 2.68 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr6_+_54249817 | 2.63 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr15_-_58828321 | 2.61 |
ENSMUST00000228067.2
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr2_-_64806106 | 2.58 |
ENSMUST00000156765.2
|
Grb14
|
growth factor receptor bound protein 14 |
| chr2_+_121978156 | 2.57 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chr11_-_35871300 | 2.57 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr10_-_53952686 | 2.47 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr1_+_171041583 | 2.43 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_-_20329823 | 2.43 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr1_+_171041539 | 2.38 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_162694076 | 2.37 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr1_+_172525613 | 2.33 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr8_+_45960804 | 2.32 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_+_124470053 | 2.29 |
ENSMUST00000049124.10
|
C1rl
|
complement component 1, r subcomponent-like |
| chr5_+_31011140 | 2.25 |
ENSMUST00000202501.2
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr6_+_91661034 | 2.15 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chrX_+_138464065 | 2.05 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr6_-_48422307 | 2.02 |
ENSMUST00000114563.8
ENSMUST00000114558.8 ENSMUST00000101443.10 |
Zfp467
|
zinc finger protein 467 |
| chr11_+_109376432 | 2.02 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr11_+_86375441 | 2.02 |
ENSMUST00000020827.7
|
Rnft1
|
ring finger protein, transmembrane 1 |
| chr16_-_11727262 | 2.01 |
ENSMUST00000127972.8
ENSMUST00000121750.2 ENSMUST00000096272.11 ENSMUST00000073371.7 |
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr7_-_126873219 | 1.98 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr16_-_20534851 | 1.97 |
ENSMUST00000120099.8
ENSMUST00000232309.2 ENSMUST00000007207.15 |
Clcn2
|
chloride channel, voltage-sensitive 2 |
| chr6_+_115398996 | 1.96 |
ENSMUST00000000450.5
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr8_+_77628916 | 1.95 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr6_+_134807097 | 1.93 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr7_+_119499322 | 1.92 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr8_+_45960931 | 1.92 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr4_-_148021159 | 1.90 |
ENSMUST00000105712.2
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr9_+_51959534 | 1.90 |
ENSMUST00000061352.11
|
Rdx
|
radixin |
| chr1_-_93373145 | 1.86 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr12_+_37291632 | 1.85 |
ENSMUST00000049874.14
|
Agmo
|
alkylglycerol monooxygenase |
| chrX_-_8059597 | 1.84 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr9_+_88209250 | 1.82 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr6_+_37847721 | 1.79 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr10_-_43880353 | 1.78 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr2_+_155359868 | 1.78 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr13_+_25127127 | 1.77 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr15_-_77813123 | 1.77 |
ENSMUST00000109748.9
ENSMUST00000109747.9 ENSMUST00000100486.6 ENSMUST00000005487.12 |
Txn2
|
thioredoxin 2 |
| chr12_+_37292029 | 1.75 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr1_+_133173826 | 1.74 |
ENSMUST00000105082.9
ENSMUST00000038295.15 |
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr8_-_122379631 | 1.74 |
ENSMUST00000046386.5
|
Zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr11_-_83421333 | 1.73 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr17_+_37269468 | 1.69 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr8_-_37200051 | 1.66 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr12_+_84332006 | 1.66 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr10_-_95251145 | 1.65 |
ENSMUST00000119917.2
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr1_+_171052623 | 1.64 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr10_-_95251327 | 1.63 |
ENSMUST00000172070.8
ENSMUST00000150432.8 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr14_+_3575879 | 1.61 |
ENSMUST00000150727.8
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr3_-_75177378 | 1.58 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr6_+_21986445 | 1.58 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_+_3575406 | 1.58 |
ENSMUST00000124353.2
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr9_+_51959415 | 1.51 |
ENSMUST00000000590.16
ENSMUST00000238858.2 |
Rdx
|
radixin |
| chr9_+_107456086 | 1.50 |
ENSMUST00000149638.8
ENSMUST00000139274.8 ENSMUST00000130053.8 ENSMUST00000122985.8 ENSMUST00000127380.8 ENSMUST00000139581.2 |
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr12_-_80483343 | 1.49 |
ENSMUST00000054145.8
|
Dcaf5
|
DDB1 and CUL4 associated factor 5 |
| chr15_-_44651411 | 1.48 |
ENSMUST00000090057.6
ENSMUST00000110269.8 ENSMUST00000228639.2 |
Sybu
|
syntabulin (syntaxin-interacting) |
| chr17_+_37269513 | 1.47 |
ENSMUST00000173814.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr15_-_99717956 | 1.42 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr6_-_93769426 | 1.40 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr16_-_56537545 | 1.36 |
ENSMUST00000141404.3
|
Tfg
|
Trk-fused gene |
| chr11_+_49500090 | 1.36 |
ENSMUST00000020617.3
|
Flt4
|
FMS-like tyrosine kinase 4 |
| chr16_-_56537650 | 1.34 |
ENSMUST00000128551.8
|
Tfg
|
Trk-fused gene |
| chr1_-_170695328 | 1.34 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chr9_-_44920899 | 1.33 |
ENSMUST00000102832.3
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr10_-_105410280 | 1.32 |
ENSMUST00000061506.9
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr14_+_67470884 | 1.32 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr14_+_5894220 | 1.32 |
ENSMUST00000063750.8
|
Rarb
|
retinoic acid receptor, beta |
| chr3_+_85946145 | 1.30 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr4_-_116565509 | 1.30 |
ENSMUST00000030453.5
|
Mmachc
|
methylmalonic aciduria cblC type, with homocystinuria |
| chr10_+_21869776 | 1.29 |
ENSMUST00000092673.11
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr14_-_30645711 | 1.28 |
ENSMUST00000006697.17
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr13_+_41013230 | 1.27 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr12_+_52550775 | 1.27 |
ENSMUST00000219443.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr6_+_108637816 | 1.26 |
ENSMUST00000163617.2
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr11_+_48977852 | 1.25 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr7_+_127311881 | 1.24 |
ENSMUST00000047393.7
|
Ctf1
|
cardiotrophin 1 |
| chr15_-_91075933 | 1.23 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr18_+_67338437 | 1.22 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr14_+_67470735 | 1.21 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr6_-_142647944 | 1.21 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr6_+_21985902 | 1.20 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr5_-_105491795 | 1.19 |
ENSMUST00000171587.2
|
Gbp11
|
guanylate binding protein 11 |
| chr19_+_56414114 | 1.17 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr7_-_37927399 | 1.17 |
ENSMUST00000098513.6
|
Plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr11_-_80030735 | 1.17 |
ENSMUST00000136996.2
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr9_+_7445822 | 1.17 |
ENSMUST00000034497.8
|
Mmp3
|
matrix metallopeptidase 3 |
| chr16_-_62667307 | 1.15 |
ENSMUST00000232561.2
ENSMUST00000089289.6 |
Arl13b
|
ADP-ribosylation factor-like 13B |
| chr16_+_90017634 | 1.15 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr17_+_43671314 | 1.14 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr8_+_25507227 | 1.14 |
ENSMUST00000033961.7
ENSMUST00000210536.2 |
Tm2d2
|
TM2 domain containing 2 |
| chr4_-_82423985 | 1.14 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr11_+_48977888 | 1.14 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr7_-_126073994 | 1.14 |
ENSMUST00000205733.2
ENSMUST00000205889.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr11_-_74228504 | 1.13 |
ENSMUST00000213831.2
|
Olfr410
|
olfactory receptor 410 |
| chr16_-_43959993 | 1.13 |
ENSMUST00000137557.8
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr2_+_113271409 | 1.12 |
ENSMUST00000081349.9
|
Fmn1
|
formin 1 |
| chr4_+_84802513 | 1.12 |
ENSMUST00000047023.13
|
Cntln
|
centlein, centrosomal protein |
| chr13_-_104056803 | 1.12 |
ENSMUST00000091269.11
ENSMUST00000188997.7 ENSMUST00000169083.8 ENSMUST00000191275.7 |
Erbin
|
Erbb2 interacting protein |
| chr12_+_76451177 | 1.11 |
ENSMUST00000219555.2
|
Hspa2
|
heat shock protein 2 |
| chrX_-_47602395 | 1.10 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr7_-_5128936 | 1.09 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr8_+_48275178 | 1.09 |
ENSMUST00000079639.3
|
Cldn24
|
claudin 24 |
| chr1_+_194302123 | 1.08 |
ENSMUST00000027952.12
|
Plxna2
|
plexin A2 |
| chr1_-_16689660 | 1.08 |
ENSMUST00000117146.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr19_-_6947076 | 1.08 |
ENSMUST00000237074.2
|
Plcb3
|
phospholipase C, beta 3 |
| chr14_-_72840373 | 1.06 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr1_+_186699613 | 1.06 |
ENSMUST00000045108.2
|
D1Pas1
|
DNA segment, Chr 1, Pasteur Institute 1 |
| chr17_+_36152559 | 1.05 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr19_+_24853039 | 1.04 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr9_-_50639230 | 1.04 |
ENSMUST00000118707.2
ENSMUST00000034566.15 |
Dixdc1
|
DIX domain containing 1 |
| chr16_-_28748410 | 1.03 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr16_+_17149235 | 1.02 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr5_+_102629240 | 1.02 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr16_-_43959559 | 1.00 |
ENSMUST00000063661.13
ENSMUST00000114666.9 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr12_+_76450941 | 1.00 |
ENSMUST00000080449.7
|
Hspa2
|
heat shock protein 2 |
| chr4_-_82423944 | 0.99 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr7_-_46445305 | 0.99 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr14_-_30645503 | 0.98 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr18_+_11052458 | 0.98 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr6_-_142647985 | 0.97 |
ENSMUST00000205202.3
ENSMUST00000073173.12 ENSMUST00000111771.8 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr2_+_23046381 | 0.97 |
ENSMUST00000028117.4
|
Yme1l1
|
YME1-like 1 (S. cerevisiae) |
| chr9_+_67747668 | 0.97 |
ENSMUST00000077879.7
|
Vps13c
|
vacuolar protein sorting 13C |
| chr3_-_88368489 | 0.96 |
ENSMUST00000166237.8
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr10_-_57408512 | 0.96 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr1_+_171265103 | 0.95 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr7_-_70010341 | 0.95 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr3_+_89987749 | 0.95 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr19_+_57349112 | 0.94 |
ENSMUST00000036407.6
|
Fam160b1
|
family with sequence similarity 160, member B1 |
| chr18_-_15536747 | 0.93 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr7_-_43956326 | 0.93 |
ENSMUST00000004587.11
|
Clec11a
|
C-type lectin domain family 11, member a |
| chr10_-_12689345 | 0.91 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr13_-_114524586 | 0.90 |
ENSMUST00000232101.2
ENSMUST00000225978.3 |
Ndufs4
|
NADH:ubiquinone oxidoreductase core subunit S4 |
| chr12_+_10440755 | 0.90 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr10_-_118705029 | 0.89 |
ENSMUST00000004281.10
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr11_-_62349334 | 0.89 |
ENSMUST00000141447.2
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr13_-_104057016 | 0.89 |
ENSMUST00000022222.12
|
Erbin
|
Erbb2 interacting protein |
| chr16_-_56537808 | 0.88 |
ENSMUST00000065515.14
|
Tfg
|
Trk-fused gene |
| chr10_+_59715439 | 0.88 |
ENSMUST00000142819.8
ENSMUST00000020309.7 |
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr12_+_76580386 | 0.88 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr7_-_89176294 | 0.87 |
ENSMUST00000207932.2
|
Prss23
|
protease, serine 23 |
| chr6_-_48818006 | 0.86 |
ENSMUST00000203229.3
|
Tmem176b
|
transmembrane protein 176B |
| chr1_+_139349912 | 0.86 |
ENSMUST00000200243.5
ENSMUST00000039867.10 |
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr6_+_134807170 | 0.85 |
ENSMUST00000111937.2
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr6_-_48818044 | 0.85 |
ENSMUST00000101429.11
ENSMUST00000204073.3 |
Tmem176b
|
transmembrane protein 176B |
| chr7_+_30157704 | 0.85 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr6_-_48818430 | 0.85 |
ENSMUST00000205147.3
|
Tmem176b
|
transmembrane protein 176B |
| chr17_+_6156738 | 0.84 |
ENSMUST00000142030.8
|
Tulp4
|
tubby like protein 4 |
| chrX_+_21581135 | 0.83 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr5_+_117501557 | 0.83 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr5_-_88823049 | 0.82 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 2.6 | 15.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 2.1 | 6.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.7 | 5.0 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 1.6 | 1.6 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 1.6 | 30.9 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 1.4 | 5.5 | GO:0030091 | protein repair(GO:0030091) |
| 1.3 | 3.8 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.0 | 9.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.0 | 3.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.9 | 2.8 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.9 | 1.8 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.9 | 2.6 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.9 | 2.6 | GO:1903989 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.8 | 5.7 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.7 | 2.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.7 | 8.0 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.7 | 17.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.7 | 2.1 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 0.7 | 6.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.7 | 0.7 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.6 | 3.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.6 | 8.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.6 | 9.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 8.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.6 | 1.8 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.6 | 3.5 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.6 | 4.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.6 | 2.2 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.5 | 4.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 2.1 | GO:2000795 | lung ciliated cell differentiation(GO:0061141) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 2.0 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.5 | 2.4 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.5 | 1.9 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.5 | 6.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 1.8 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.5 | 0.5 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.4 | 14.9 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.4 | 2.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.4 | 1.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.4 | 5.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.4 | 3.4 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.4 | 3.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 1.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.3 | 2.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 2.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 1.8 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.3 | 2.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 2.0 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.3 | 0.8 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.3 | 3.0 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.3 | 1.8 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.3 | 2.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 0.8 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.2 | 1.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 1.0 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 | 1.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 1.6 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 0.4 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.2 | 1.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 0.9 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.2 | 1.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.4 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 1.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.2 | 0.6 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 1.0 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.2 | 1.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.9 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 1.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 2.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 1.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.2 | 1.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.2 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.2 | 1.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 1.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 0.6 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.2 | 1.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 2.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.3 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.9 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 1.2 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 1.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.1 | 0.5 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) |
| 0.1 | 3.9 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.1 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 0.6 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.5 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 4.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.5 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 1.4 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 2.7 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.1 | 3.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 1.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.6 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 2.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 9.6 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 0.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.2 | GO:0034442 | regulation of lipoprotein oxidation(GO:0034442) |
| 0.1 | 1.1 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 | 0.9 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 4.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.3 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 1.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 2.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.2 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.1 | 2.6 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.2 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.2 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 2.0 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.2 | GO:0060618 | nipple development(GO:0060618) |
| 0.1 | 1.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.3 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.1 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.6 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.7 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 3.0 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.1 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 0.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.7 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:0061296 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) pronephros morphogenesis(GO:0072114) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.0 | 2.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 1.3 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.4 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 1.7 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 2.8 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.5 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 1.3 | GO:0043153 | photoperiodism(GO:0009648) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.6 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.2 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 4.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.5 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 2.2 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 1.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 1.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 1.2 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) |
| 0.0 | 1.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 4.2 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.9 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.6 | 5.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 3.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.4 | 6.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 1.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.3 | 2.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 3.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.3 | 2.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 1.5 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 2.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 1.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 4.6 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 7.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 2.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 5.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 16.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 3.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 2.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 3.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 3.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.0 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 4.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 59.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.0 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 21.1 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 17.9 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 3.1 | 15.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 2.4 | 9.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 2.1 | 6.3 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 2.0 | 9.9 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 1.7 | 5.0 | GO:0005369 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 1.5 | 30.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 1.3 | 8.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.2 | 6.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 1.2 | 7.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.2 | 3.5 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.0 | 3.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.9 | 2.8 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.8 | 2.4 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.8 | 3.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.7 | 30.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.7 | 8.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.7 | 2.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.6 | 1.9 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.6 | 7.9 | GO:0035473 | lipase binding(GO:0035473) |
| 0.6 | 1.7 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 3.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 9.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 3.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 2.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.5 | 2.8 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.4 | 1.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 1.7 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.4 | 1.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 1.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.4 | 4.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.4 | 1.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.3 | 4.8 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 1.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 1.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 1.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.3 | 3.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 0.8 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.3 | 17.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.3 | 1.8 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 0.7 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 1.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 2.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 6.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.8 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 2.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 1.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 4.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.9 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 2.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 0.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 6.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.7 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.1 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.8 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 2.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 3.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 2.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 2.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 1.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.8 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.1 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.5 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.9 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 4.7 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 0.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 5.5 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 2.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 4.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.7 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 3.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 3.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 15.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 16.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 1.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 5.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 4.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 3.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 3.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 4.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 4.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 2.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.7 | 34.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.6 | 18.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.5 | 17.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.5 | 9.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 4.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 9.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 9.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 2.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 5.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 2.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 19.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 3.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 8.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 3.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 4.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 3.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |