Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
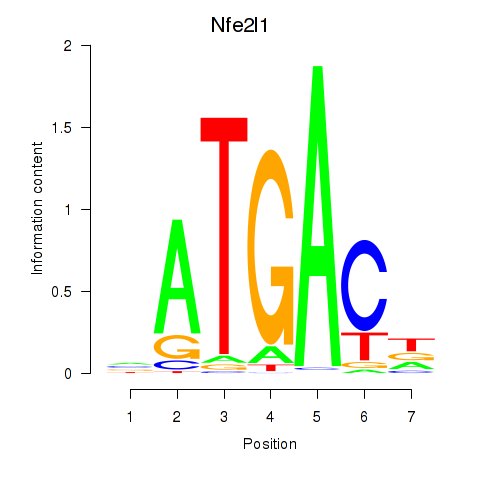
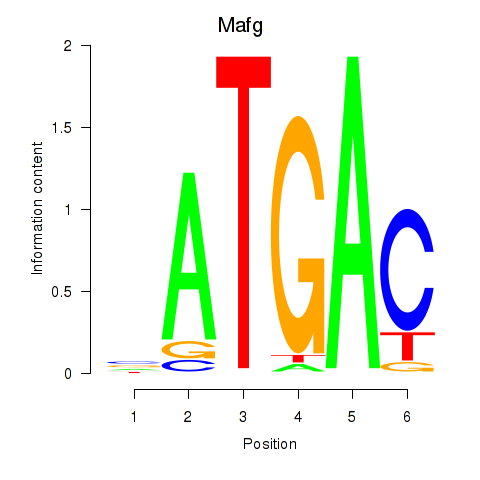
Results for Nfe2l1_Mafg
Z-value: 0.31
Transcription factors associated with Nfe2l1_Mafg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l1
|
ENSMUSG00000038615.18 | nuclear factor, erythroid derived 2,-like 1 |
|
Mafg
|
ENSMUSG00000051510.14 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2l1 | mm39_v1_chr11_-_96720309_96720328 | -0.40 | 1.6e-02 | Click! |
| Mafg | mm39_v1_chr11_-_120524362_120524431 | 0.19 | 2.7e-01 | Click! |
Activity profile of Nfe2l1_Mafg motif
Sorted Z-values of Nfe2l1_Mafg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_78563513 | 1.30 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr1_-_66984178 | 1.24 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr2_+_130119077 | 0.99 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr11_+_106916430 | 0.96 |
ENSMUST00000140447.4
|
1810010H24Rik
|
RIKEN cDNA 1810010H24 gene |
| chr1_-_66984521 | 0.93 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chrX_-_48877080 | 0.74 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr13_-_55677109 | 0.74 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3 |
| chr18_-_34505544 | 0.67 |
ENSMUST00000236887.2
|
Reep5
|
receptor accessory protein 5 |
| chr4_+_126915104 | 0.63 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr8_+_95710977 | 0.57 |
ENSMUST00000093271.8
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr14_-_70855980 | 0.51 |
ENSMUST00000228001.2
|
Dmtn
|
dematin actin binding protein |
| chr12_+_17398421 | 0.50 |
ENSMUST00000046011.12
|
Nol10
|
nucleolar protein 10 |
| chr17_-_48739874 | 0.49 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr12_-_55033130 | 0.48 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr15_-_101833160 | 0.48 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr6_+_142244145 | 0.47 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr4_-_131776368 | 0.45 |
ENSMUST00000105981.9
ENSMUST00000084253.10 ENSMUST00000141291.3 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr19_+_38121248 | 0.44 |
ENSMUST00000025956.13
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr6_+_41498716 | 0.44 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr19_+_38121214 | 0.43 |
ENSMUST00000112329.3
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr1_-_133681419 | 0.39 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr17_+_36132567 | 0.39 |
ENSMUST00000003635.7
|
Ier3
|
immediate early response 3 |
| chr11_+_48691175 | 0.38 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr6_-_86646118 | 0.38 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr13_-_55676334 | 0.38 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr7_-_16651107 | 0.37 |
ENSMUST00000173139.2
|
Calm3
|
calmodulin 3 |
| chr14_-_58063585 | 0.36 |
ENSMUST00000022536.3
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr9_+_108437485 | 0.36 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr3_+_157272504 | 0.35 |
ENSMUST00000041175.13
ENSMUST00000173533.2 |
Ptger3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr2_-_25162347 | 0.35 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr11_-_120238917 | 0.34 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr7_-_115637970 | 0.34 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr18_-_43610829 | 0.33 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr5_-_65548723 | 0.33 |
ENSMUST00000120094.5
ENSMUST00000127874.6 ENSMUST00000057885.13 |
Rpl9
|
ribosomal protein L9 |
| chrX_+_55500170 | 0.32 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr10_+_80765900 | 0.32 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr10_+_82821304 | 0.31 |
ENSMUST00000040110.8
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chrX_+_70600481 | 0.30 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr13_-_3854307 | 0.30 |
ENSMUST00000077698.5
|
Calml3
|
calmodulin-like 3 |
| chr3_-_103698853 | 0.29 |
ENSMUST00000118317.8
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr15_-_101602734 | 0.29 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr8_+_95711037 | 0.29 |
ENSMUST00000211944.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr5_-_124170305 | 0.28 |
ENSMUST00000040967.9
|
Vps37b
|
vacuolar protein sorting 37B |
| chr10_-_13744676 | 0.28 |
ENSMUST00000019942.6
ENSMUST00000162610.8 |
Aig1
|
androgen-induced 1 |
| chr1_-_82746169 | 0.28 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr14_+_74878280 | 0.27 |
ENSMUST00000036653.5
|
Htr2a
|
5-hydroxytryptamine (serotonin) receptor 2A |
| chr4_-_63321591 | 0.26 |
ENSMUST00000035724.5
|
Akna
|
AT-hook transcription factor |
| chr10_-_13744523 | 0.25 |
ENSMUST00000105534.10
|
Aig1
|
androgen-induced 1 |
| chr12_-_31684588 | 0.25 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr1_+_82817388 | 0.25 |
ENSMUST00000190052.7
ENSMUST00000063380.11 ENSMUST00000187899.7 ENSMUST00000186302.7 ENSMUST00000190046.7 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr3_-_82957104 | 0.22 |
ENSMUST00000048246.5
|
Fgb
|
fibrinogen beta chain |
| chr15_+_79575046 | 0.22 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr4_+_132262853 | 0.22 |
ENSMUST00000094657.10
ENSMUST00000105940.10 ENSMUST00000105939.10 ENSMUST00000150207.8 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr14_-_70945434 | 0.22 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr17_-_71305003 | 0.22 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr9_-_123778334 | 0.22 |
ENSMUST00000071404.5
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr11_+_51510555 | 0.22 |
ENSMUST00000127405.2
|
Nhp2
|
NHP2 ribonucleoprotein |
| chr5_+_145060489 | 0.21 |
ENSMUST00000136074.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr1_+_82817170 | 0.21 |
ENSMUST00000189220.7
ENSMUST00000113444.8 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr11_-_69556904 | 0.21 |
ENSMUST00000018918.12
|
Cd68
|
CD68 antigen |
| chr2_+_119572770 | 0.20 |
ENSMUST00000028758.8
|
Itpka
|
inositol 1,4,5-trisphosphate 3-kinase A |
| chr3_-_103698391 | 0.20 |
ENSMUST00000106845.9
ENSMUST00000029438.15 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr2_-_121101822 | 0.19 |
ENSMUST00000110647.8
ENSMUST00000110648.8 |
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr15_-_102097387 | 0.19 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr3_+_88523440 | 0.19 |
ENSMUST00000177498.8
ENSMUST00000176500.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr5_+_31855394 | 0.16 |
ENSMUST00000063813.11
ENSMUST00000071531.12 ENSMUST00000131995.7 ENSMUST00000114507.10 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr10_-_75946790 | 0.15 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr15_-_102097466 | 0.15 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr11_-_109363406 | 0.14 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr11_-_116743898 | 0.14 |
ENSMUST00000190993.7
ENSMUST00000092404.13 |
Srsf2
|
serine and arginine-rich splicing factor 2 |
| chr4_-_41275091 | 0.14 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr17_+_8144822 | 0.14 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr19_-_5416339 | 0.14 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr6_+_40941688 | 0.14 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr7_+_127172701 | 0.14 |
ENSMUST00000121004.3
|
Phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr5_+_31070739 | 0.13 |
ENSMUST00000031055.8
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr8_+_125624615 | 0.13 |
ENSMUST00000034467.7
|
Sprtn
|
SprT-like N-terminal domain |
| chr9_+_21279179 | 0.13 |
ENSMUST00000213518.2
ENSMUST00000216892.2 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr1_+_45834645 | 0.13 |
ENSMUST00000147308.2
|
Wdr75
|
WD repeat domain 75 |
| chr5_+_31855304 | 0.13 |
ENSMUST00000114515.9
|
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr11_-_51891575 | 0.13 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chrX_+_20554618 | 0.13 |
ENSMUST00000033380.7
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr12_+_69418886 | 0.13 |
ENSMUST00000050063.9
|
Arf6
|
ADP-ribosylation factor 6 |
| chr11_+_116324913 | 0.13 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr9_+_111011388 | 0.13 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr9_+_21279299 | 0.12 |
ENSMUST00000214852.2
ENSMUST00000115414.3 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr1_+_127701901 | 0.12 |
ENSMUST00000112570.2
ENSMUST00000027587.15 |
Ccnt2
|
cyclin T2 |
| chr2_+_120439858 | 0.12 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr12_-_112964279 | 0.12 |
ENSMUST00000011302.9
|
Brf1
|
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr1_+_82817794 | 0.12 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr12_-_56392646 | 0.12 |
ENSMUST00000021416.9
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chrX_-_153999440 | 0.12 |
ENSMUST00000026318.15
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr3_+_102965601 | 0.12 |
ENSMUST00000029445.13
ENSMUST00000200069.5 |
Nras
|
neuroblastoma ras oncogene |
| chr11_-_69556888 | 0.11 |
ENSMUST00000108654.3
|
Cd68
|
CD68 antigen |
| chr18_-_47466378 | 0.11 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr15_+_44059531 | 0.11 |
ENSMUST00000038856.14
ENSMUST00000110289.4 |
Trhr
|
thyrotropin releasing hormone receptor |
| chr17_+_31329939 | 0.11 |
ENSMUST00000236241.2
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr3_+_135144481 | 0.11 |
ENSMUST00000199582.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_-_103304573 | 0.11 |
ENSMUST00000149431.2
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr4_+_101365052 | 0.11 |
ENSMUST00000038207.12
|
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr9_+_21279161 | 0.10 |
ENSMUST00000067646.12
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr8_-_76133212 | 0.10 |
ENSMUST00000212864.2
|
Gm10358
|
predicted gene 10358 |
| chr3_+_88523730 | 0.10 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr5_-_107873883 | 0.10 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr12_-_46863726 | 0.10 |
ENSMUST00000219330.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr12_+_38833501 | 0.10 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chr1_-_131127825 | 0.09 |
ENSMUST00000068564.15
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr8_-_40964704 | 0.09 |
ENSMUST00000135269.8
ENSMUST00000034012.10 |
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr12_+_85017671 | 0.09 |
ENSMUST00000021669.15
ENSMUST00000171040.2 |
Fcf1
|
FCF1 rRNA processing protein |
| chr12_+_38833454 | 0.09 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chr3_+_88536749 | 0.09 |
ENSMUST00000176316.8
ENSMUST00000176879.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr9_-_122942196 | 0.09 |
ENSMUST00000238520.2
ENSMUST00000147563.9 |
Zdhhc3
|
zinc finger, DHHC domain containing 3 |
| chr5_-_65248962 | 0.09 |
ENSMUST00000212080.2
|
Tmem156
|
transmembrane protein 156 |
| chr1_-_182929025 | 0.09 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr19_-_4861536 | 0.09 |
ENSMUST00000179909.8
ENSMUST00000172000.3 ENSMUST00000180008.2 ENSMUST00000113793.4 ENSMUST00000006625.8 |
Gm21992
Rbm14
|
predicted gene 21992 RNA binding motif protein 14 |
| chr14_-_52273600 | 0.09 |
ENSMUST00000214342.2
|
Olfr221
|
olfactory receptor 221 |
| chr2_-_30793266 | 0.08 |
ENSMUST00000102852.6
|
Ptges
|
prostaglandin E synthase |
| chr13_+_43276323 | 0.08 |
ENSMUST00000136576.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr14_+_79718604 | 0.08 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1 |
| chr2_+_112092271 | 0.08 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr11_+_78717398 | 0.08 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr15_-_101389384 | 0.08 |
ENSMUST00000023718.9
|
Krt83
|
keratin 83 |
| chr8_-_40964282 | 0.08 |
ENSMUST00000149992.2
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr17_+_71511642 | 0.08 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr14_-_122202599 | 0.08 |
ENSMUST00000049872.9
|
Gpr183
|
G protein-coupled receptor 183 |
| chr18_+_52958382 | 0.08 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr5_-_65248927 | 0.08 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr8_+_66838927 | 0.07 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr2_-_76700830 | 0.07 |
ENSMUST00000138542.2
|
Ttn
|
titin |
| chr6_+_15196950 | 0.07 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr9_-_45923908 | 0.07 |
ENSMUST00000217514.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr15_-_80989200 | 0.07 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr7_+_101546059 | 0.07 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr6_-_129484070 | 0.07 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr8_+_117983803 | 0.07 |
ENSMUST00000166750.9
|
Cmip
|
c-Maf inducing protein |
| chr4_-_129534403 | 0.07 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr10_+_51898598 | 0.07 |
ENSMUST00000163017.10
|
Vgll2
|
vestigial like family member 2 |
| chr17_-_33979442 | 0.07 |
ENSMUST00000057373.14
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr11_+_115824108 | 0.06 |
ENSMUST00000140991.2
|
Sap30bp
|
SAP30 binding protein |
| chr19_-_40982576 | 0.06 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr15_-_41733099 | 0.06 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr15_-_42540363 | 0.06 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr6_-_68857658 | 0.06 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr1_+_87332638 | 0.06 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr10_-_23977810 | 0.06 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr4_-_132611820 | 0.06 |
ENSMUST00000030698.5
|
Stx12
|
syntaxin 12 |
| chr2_+_25162487 | 0.06 |
ENSMUST00000028341.11
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr1_-_84818223 | 0.06 |
ENSMUST00000186465.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr9_+_53678801 | 0.05 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr9_-_83028654 | 0.05 |
ENSMUST00000161796.9
ENSMUST00000162246.9 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr3_-_144514386 | 0.05 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr8_+_83891972 | 0.05 |
ENSMUST00000034145.11
|
Tbc1d9
|
TBC1 domain family, member 9 |
| chr8_-_73228953 | 0.05 |
ENSMUST00000079510.6
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr7_+_30475819 | 0.05 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr4_-_123507494 | 0.05 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr7_-_102506478 | 0.05 |
ENSMUST00000071393.3
|
Olfr566
|
olfactory receptor 566 |
| chr8_-_37420293 | 0.05 |
ENSMUST00000179501.2
|
Dlc1
|
deleted in liver cancer 1 |
| chr6_+_92793440 | 0.05 |
ENSMUST00000057977.4
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr2_+_128809268 | 0.05 |
ENSMUST00000110320.9
ENSMUST00000110319.3 |
Zc3h6
|
zinc finger CCCH type containing 6 |
| chr3_-_104960437 | 0.05 |
ENSMUST00000077548.12
|
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr3_-_104960264 | 0.04 |
ENSMUST00000098763.7
ENSMUST00000197437.5 |
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr1_+_31215482 | 0.04 |
ENSMUST00000062560.14
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr5_-_51711237 | 0.04 |
ENSMUST00000132734.8
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr6_-_12749409 | 0.04 |
ENSMUST00000119581.7
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr19_-_11291805 | 0.04 |
ENSMUST00000187467.2
|
Ms4a14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr1_+_172784335 | 0.04 |
ENSMUST00000038432.7
|
Olfr16
|
olfactory receptor 16 |
| chr1_+_173986288 | 0.04 |
ENSMUST00000068403.4
|
Olfr420
|
olfactory receptor 420 |
| chr8_-_36293588 | 0.04 |
ENSMUST00000060128.7
|
Cldn23
|
claudin 23 |
| chr14_+_58063668 | 0.04 |
ENSMUST00000022538.5
|
Mrpl57
|
mitochondrial ribosomal protein L57 |
| chr11_-_50183129 | 0.04 |
ENSMUST00000059458.5
|
Maml1
|
mastermind like transcriptional coactivator 1 |
| chr9_+_23285228 | 0.04 |
ENSMUST00000214050.2
|
Bmper
|
BMP-binding endothelial regulator |
| chr12_+_84408742 | 0.04 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr3_+_7494108 | 0.04 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr5_-_136227760 | 0.04 |
ENSMUST00000149151.2
ENSMUST00000151786.8 |
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chr6_+_11926757 | 0.04 |
ENSMUST00000133776.2
|
Phf14
|
PHD finger protein 14 |
| chr5_+_24679154 | 0.04 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr11_-_85125889 | 0.04 |
ENSMUST00000018625.10
|
Appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr19_-_5781503 | 0.04 |
ENSMUST00000162976.2
|
Znrd2
|
zinc ribbon domain containing 2 |
| chr10_+_112292161 | 0.03 |
ENSMUST00000219607.2
ENSMUST00000218827.2 |
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr8_-_70389147 | 0.03 |
ENSMUST00000212277.2
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr6_-_98319684 | 0.03 |
ENSMUST00000164491.3
|
Mdfic2
|
MyoD family inhibitor domain containing 2 |
| chr2_-_148250024 | 0.03 |
ENSMUST00000099270.5
|
Thbd
|
thrombomodulin |
| chr5_-_51711204 | 0.03 |
ENSMUST00000196968.5
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr11_+_86574811 | 0.03 |
ENSMUST00000108022.8
ENSMUST00000108021.2 |
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr13_+_111391544 | 0.03 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr3_+_89970088 | 0.03 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr7_+_103578032 | 0.03 |
ENSMUST00000106863.2
|
Olfr631
|
olfactory receptor 631 |
| chr9_-_66741998 | 0.03 |
ENSMUST00000169282.8
|
Aph1c
|
aph1 homolog C, gamma secretase subunit |
| chr5_+_35074862 | 0.03 |
ENSMUST00000202205.2
|
Msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr1_-_181985663 | 0.03 |
ENSMUST00000169123.4
|
Vmn1r1
|
vomeronasal 1 receptor 1 |
| chr4_+_109263820 | 0.02 |
ENSMUST00000124209.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr3_-_92407800 | 0.02 |
ENSMUST00000062129.2
|
Sprr4
|
small proline-rich protein 4 |
| chr10_-_129962924 | 0.02 |
ENSMUST00000074161.2
|
Olfr824
|
olfactory receptor 824 |
| chr11_-_120441952 | 0.02 |
ENSMUST00000026121.3
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr9_-_15834052 | 0.02 |
ENSMUST00000217187.2
|
Fat3
|
FAT atypical cadherin 3 |
| chr3_-_37286714 | 0.02 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr2_+_86128161 | 0.02 |
ENSMUST00000054746.5
|
Olfr1052
|
olfactory receptor 1052 |
| chr3_+_40978804 | 0.02 |
ENSMUST00000099121.10
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr1_-_97689263 | 0.02 |
ENSMUST00000171129.8
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr11_-_97657327 | 0.02 |
ENSMUST00000107579.2
ENSMUST00000018685.9 |
Cwc25
|
CWC25 spliceosome-associated protein |
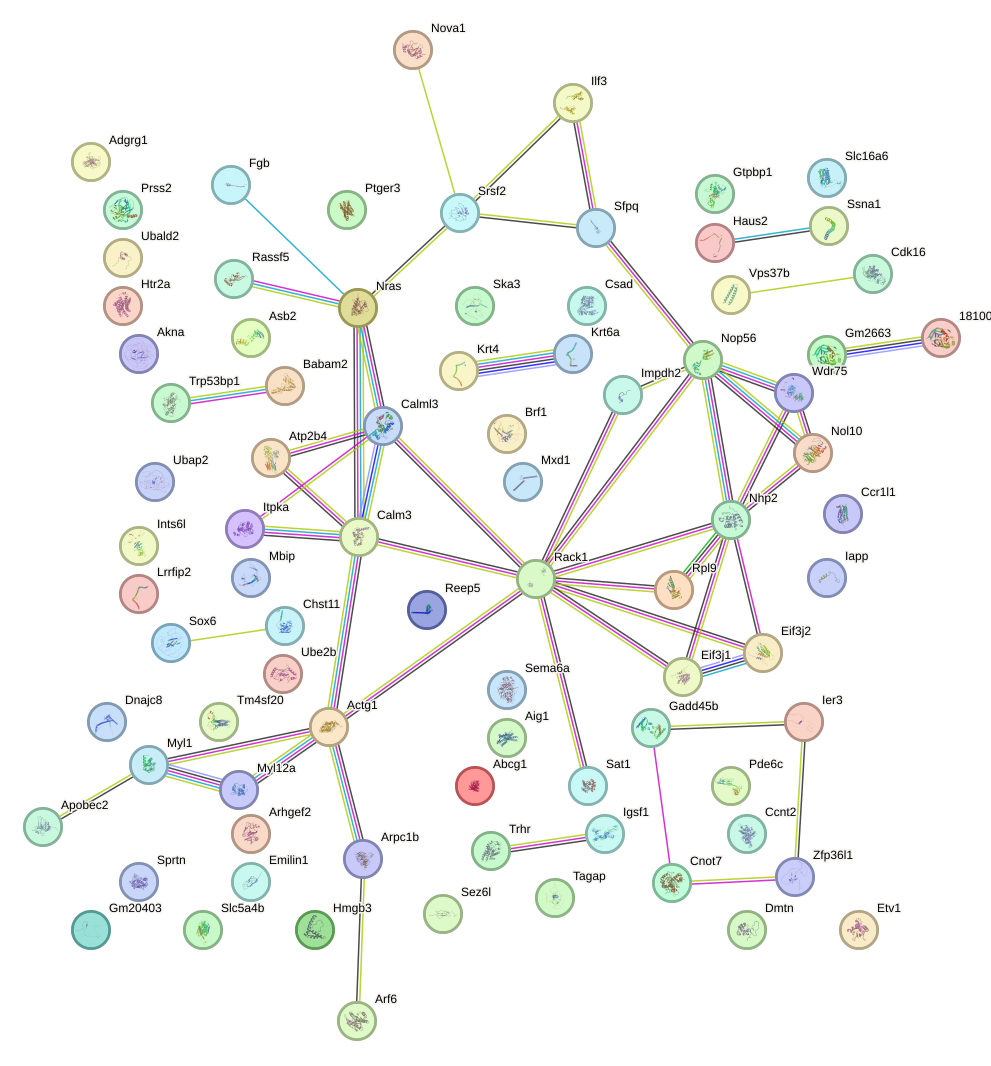
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l1_Mafg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.4 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.4 | GO:1990764 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of actin filament-based movement(GO:1903116) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 0.4 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 0.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.5 | GO:1901731 | calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.3 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.5 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.6 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.3 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.0 | 0.5 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.0 | 0.1 | GO:0070103 | positive regulation of cell fate specification(GO:0042660) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:1904633 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.0 | 0.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0055099 | glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0030210 | negative regulation of cytokine secretion involved in immune response(GO:0002740) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0043545 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.3 | GO:0044146 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.1 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 1.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.9 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.3 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 2.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.4 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.9 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.4 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.0 | 1.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |