Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
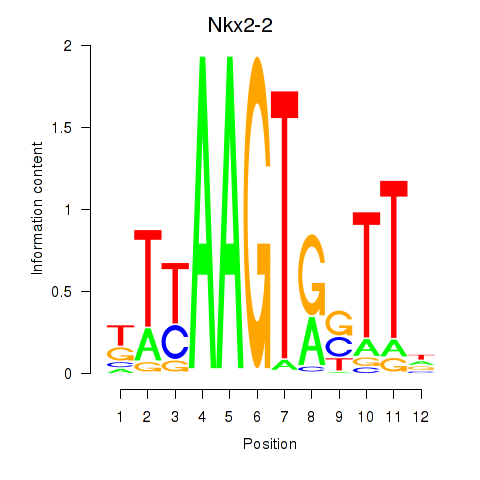
Results for Nkx2-2
Z-value: 0.86
Transcription factors associated with Nkx2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-2
|
ENSMUSG00000027434.12 | NK2 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-2 | mm39_v1_chr2_-_147028309_147028331 | -0.14 | 4.2e-01 | Click! |
Activity profile of Nkx2-2 motif
Sorted Z-values of Nkx2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41681273 | 4.08 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr2_+_174292471 | 3.94 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr9_+_72345801 | 3.19 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr11_+_78192355 | 2.38 |
ENSMUST00000045026.4
|
Spag5
|
sperm associated antigen 5 |
| chr6_+_86055018 | 2.33 |
ENSMUST00000205034.3
ENSMUST00000203724.3 |
Add2
|
adducin 2 (beta) |
| chr4_+_46039202 | 2.28 |
ENSMUST00000156200.8
|
Tmod1
|
tropomodulin 1 |
| chr6_-_70769135 | 2.04 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr11_-_79971750 | 1.99 |
ENSMUST00000103233.10
ENSMUST00000061283.15 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr6_-_136834725 | 1.92 |
ENSMUST00000032341.3
|
Art4
|
ADP-ribosyltransferase 4 |
| chr1_+_40478787 | 1.92 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr6_+_86055048 | 1.89 |
ENSMUST00000032069.8
|
Add2
|
adducin 2 (beta) |
| chr7_+_110371811 | 1.88 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr17_+_18108102 | 1.81 |
ENSMUST00000054871.12
ENSMUST00000064068.5 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr17_+_43879496 | 1.68 |
ENSMUST00000169694.2
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr12_+_105996961 | 1.64 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr5_-_34326847 | 1.64 |
ENSMUST00000202042.2
ENSMUST00000060049.8 |
Haus3
|
HAUS augmin-like complex, subunit 3 |
| chr17_+_18108086 | 1.61 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr10_-_93727003 | 1.60 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr2_+_83474779 | 1.59 |
ENSMUST00000081591.7
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr11_+_23234644 | 1.58 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr19_-_60770628 | 1.58 |
ENSMUST00000238125.2
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr2_-_140012447 | 1.55 |
ENSMUST00000046030.8
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr3_-_104771716 | 1.43 |
ENSMUST00000195912.2
ENSMUST00000094028.10 |
Capza1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chr5_-_110987441 | 1.42 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr14_-_56026266 | 1.42 |
ENSMUST00000168716.8
ENSMUST00000178399.3 ENSMUST00000022830.14 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr17_+_48653493 | 1.37 |
ENSMUST00000113237.4
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr17_+_43879366 | 1.28 |
ENSMUST00000167418.8
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr5_+_110987839 | 1.28 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr5_-_110987604 | 1.27 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr5_-_69749617 | 1.26 |
ENSMUST00000173927.8
ENSMUST00000120789.8 ENSMUST00000031117.13 |
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr1_-_80255156 | 1.23 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr11_+_48691175 | 1.22 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1 |
| chr15_-_10470575 | 1.21 |
ENSMUST00000136591.8
|
Dnajc21
|
DnaJ heat shock protein family (Hsp40) member C21 |
| chrX_+_141608694 | 1.21 |
ENSMUST00000112888.2
|
Tmem164
|
transmembrane protein 164 |
| chr13_+_104315301 | 1.20 |
ENSMUST00000022225.12
ENSMUST00000069187.12 |
Trim23
|
tripartite motif-containing 23 |
| chr9_+_51124983 | 1.20 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr13_+_38220971 | 1.19 |
ENSMUST00000021866.10
|
Riok1
|
RIO kinase 1 |
| chr9_-_45896663 | 1.18 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr1_-_152262425 | 1.15 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr10_-_117681864 | 1.06 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr13_-_76166789 | 1.05 |
ENSMUST00000179078.9
ENSMUST00000167271.9 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr11_-_79414542 | 1.05 |
ENSMUST00000179322.2
|
Evi2b
|
ecotropic viral integration site 2b |
| chr3_-_130524024 | 1.05 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chrX_+_74425990 | 1.01 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr12_+_73948143 | 1.00 |
ENSMUST00000110461.8
|
Hif1a
|
hypoxia inducible factor 1, alpha subunit |
| chr13_-_21637830 | 0.99 |
ENSMUST00000032820.15
|
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr17_+_71923210 | 0.99 |
ENSMUST00000047086.10
|
Wdr43
|
WD repeat domain 43 |
| chr13_-_21637874 | 0.99 |
ENSMUST00000110485.3
|
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr15_+_79578141 | 0.98 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr14_-_72840373 | 0.97 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr3_+_10431961 | 0.96 |
ENSMUST00000029049.7
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr9_+_35470752 | 0.94 |
ENSMUST00000034615.10
ENSMUST00000121246.2 |
Pus3
|
pseudouridine synthase 3 |
| chr7_-_43182595 | 0.94 |
ENSMUST00000205503.2
|
Cd33
|
CD33 antigen |
| chr10_+_25317309 | 0.93 |
ENSMUST00000217929.2
ENSMUST00000220121.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr2_+_173918715 | 0.93 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr1_-_85664246 | 0.92 |
ENSMUST00000064788.14
|
A630001G21Rik
|
RIKEN cDNA A630001G21 gene |
| chr14_+_24540745 | 0.91 |
ENSMUST00000112384.10
|
Rps24
|
ribosomal protein S24 |
| chr13_-_113237505 | 0.91 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr14_+_24540815 | 0.88 |
ENSMUST00000224568.2
|
Rps24
|
ribosomal protein S24 |
| chr6_+_21985902 | 0.88 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr18_+_6765145 | 0.88 |
ENSMUST00000234810.2
ENSMUST00000234626.2 ENSMUST00000097680.7 |
Rab18
|
RAB18, member RAS oncogene family |
| chr14_+_24540777 | 0.87 |
ENSMUST00000169826.3
ENSMUST00000225023.2 ENSMUST00000223999.2 |
Rps24
|
ribosomal protein S24 |
| chr6_-_120334382 | 0.87 |
ENSMUST00000032283.12
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr13_-_20008397 | 0.87 |
ENSMUST00000222664.2
ENSMUST00000065335.3 |
Gpr141
|
G protein-coupled receptor 141 |
| chr6_+_8520006 | 0.86 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr8_+_124059414 | 0.86 |
ENSMUST00000010298.7
|
Spire2
|
spire type actin nucleation factor 2 |
| chr1_+_40478926 | 0.85 |
ENSMUST00000173514.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_-_152262339 | 0.84 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15 |
| chr14_-_104760051 | 0.83 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr4_-_15149755 | 0.82 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr3_-_146476331 | 0.82 |
ENSMUST00000106138.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr16_-_30152708 | 0.82 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chr4_+_124923785 | 0.81 |
ENSMUST00000030684.8
|
Gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr19_-_6065872 | 0.80 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1 |
| chr10_-_88601443 | 0.80 |
ENSMUST00000220188.2
ENSMUST00000218967.2 |
Utp20
|
UTP20 small subunit processome component |
| chrX_+_163763588 | 0.76 |
ENSMUST00000167446.8
ENSMUST00000057150.8 |
Fancb
|
Fanconi anemia, complementation group B |
| chr7_-_122969064 | 0.75 |
ENSMUST00000207010.2
ENSMUST00000167309.8 ENSMUST00000205262.2 ENSMUST00000106442.9 ENSMUST00000098060.5 |
Arhgap17
|
Rho GTPase activating protein 17 |
| chr3_-_146475974 | 0.74 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr6_+_5390386 | 0.73 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr8_+_95393349 | 0.72 |
ENSMUST00000109527.6
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr2_+_170353338 | 0.72 |
ENSMUST00000136839.2
ENSMUST00000109148.8 ENSMUST00000170167.8 |
Pfdn4
|
prefoldin 4 |
| chr10_+_22236451 | 0.71 |
ENSMUST00000182677.8
|
Raet1d
|
retinoic acid early transcript delta |
| chr1_-_173703424 | 0.71 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr12_+_33003882 | 0.71 |
ENSMUST00000076698.13
|
Sypl
|
synaptophysin-like protein |
| chr15_+_6552270 | 0.69 |
ENSMUST00000226412.2
|
Fyb
|
FYN binding protein |
| chr6_+_21986445 | 0.68 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr13_+_23940964 | 0.67 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr17_-_66191912 | 0.66 |
ENSMUST00000024905.11
|
Ralbp1
|
ralA binding protein 1 |
| chr6_-_120334400 | 0.65 |
ENSMUST00000112703.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr8_+_41692755 | 0.65 |
ENSMUST00000210862.2
ENSMUST00000045218.9 ENSMUST00000211247.2 |
Pcm1
|
pericentriolar material 1 |
| chr1_-_9818601 | 0.62 |
ENSMUST00000057438.7
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr14_+_76714350 | 0.61 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr13_-_103901010 | 0.61 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr7_-_100504610 | 0.60 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr2_+_62494622 | 0.60 |
ENSMUST00000028257.3
|
Gca
|
grancalcin |
| chr4_+_59626185 | 0.60 |
ENSMUST00000070150.11
ENSMUST00000052420.7 |
E130308A19Rik
|
RIKEN cDNA E130308A19 gene |
| chr10_-_13199971 | 0.59 |
ENSMUST00000105543.9
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr2_+_3705824 | 0.59 |
ENSMUST00000115054.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr3_-_59127571 | 0.58 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr3_-_72875187 | 0.58 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chrX_+_105070865 | 0.57 |
ENSMUST00000113557.8
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr13_+_35059285 | 0.56 |
ENSMUST00000077853.5
|
Prpf4b
|
pre-mRNA processing factor 4B |
| chr4_+_3940747 | 0.56 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chrX_+_105070907 | 0.55 |
ENSMUST00000055941.7
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr2_+_43638814 | 0.54 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr8_-_23747023 | 0.54 |
ENSMUST00000121783.7
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr2_-_58050494 | 0.53 |
ENSMUST00000028175.7
|
Cytip
|
cytohesin 1 interacting protein |
| chr8_-_23747057 | 0.52 |
ENSMUST00000051094.9
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr15_+_66542598 | 0.52 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin |
| chr5_+_76677150 | 0.52 |
ENSMUST00000087133.11
ENSMUST00000113493.8 ENSMUST00000049469.13 |
Exoc1
|
exocyst complex component 1 |
| chrX_+_67864699 | 0.51 |
ENSMUST00000096420.3
|
Gm14698
|
predicted gene 14698 |
| chr11_+_52265090 | 0.51 |
ENSMUST00000020673.3
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr7_+_120634834 | 0.51 |
ENSMUST00000207351.2
|
Mettl9
|
methyltransferase like 9 |
| chr6_+_88442391 | 0.51 |
ENSMUST00000032165.16
|
Ruvbl1
|
RuvB-like protein 1 |
| chr18_+_4993795 | 0.50 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr11_-_90281721 | 0.50 |
ENSMUST00000004051.8
|
Hlf
|
hepatic leukemia factor |
| chr19_-_11243530 | 0.49 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr13_+_47347301 | 0.49 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr11_-_45845992 | 0.49 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr11_+_75521800 | 0.49 |
ENSMUST00000006286.9
|
Inpp5k
|
inositol polyphosphate 5-phosphatase K |
| chr2_-_77776675 | 0.49 |
ENSMUST00000111821.9
ENSMUST00000111818.8 |
Cwc22
|
CWC22 spliceosome-associated protein |
| chrX_+_100317629 | 0.48 |
ENSMUST00000117706.8
|
Med12
|
mediator complex subunit 12 |
| chr14_-_54923517 | 0.47 |
ENSMUST00000125265.2
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr13_-_67080968 | 0.47 |
ENSMUST00000172597.8
ENSMUST00000173773.2 |
Mterf3
|
mitochondrial transcription termination factor 3 |
| chr7_+_46490899 | 0.46 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chrX_+_9751861 | 0.45 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr6_+_68233361 | 0.45 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr4_+_125918333 | 0.45 |
ENSMUST00000106162.8
|
Csf3r
|
colony stimulating factor 3 receptor (granulocyte) |
| chr2_-_73284262 | 0.45 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr7_-_43182504 | 0.44 |
ENSMUST00000004728.12
|
Cd33
|
CD33 antigen |
| chrX_-_100463810 | 0.44 |
ENSMUST00000118092.8
ENSMUST00000119699.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr12_-_72455708 | 0.43 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr2_+_163503415 | 0.43 |
ENSMUST00000135537.8
|
Pkig
|
protein kinase inhibitor, gamma |
| chr17_+_29833760 | 0.43 |
ENSMUST00000024817.15
ENSMUST00000162588.3 |
Rnf8
|
ring finger protein 8 |
| chr12_+_59176506 | 0.43 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr12_+_59176543 | 0.42 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr17_+_71326510 | 0.41 |
ENSMUST00000073211.13
ENSMUST00000024847.14 |
Myom1
|
myomesin 1 |
| chr9_-_119151428 | 0.40 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr2_+_173918089 | 0.40 |
ENSMUST00000155000.8
ENSMUST00000134876.8 ENSMUST00000147038.8 |
Stx16
|
syntaxin 16 |
| chr10_-_13264574 | 0.39 |
ENSMUST00000079698.7
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr18_+_23886765 | 0.38 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_+_45035942 | 0.38 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chr6_-_120334302 | 0.37 |
ENSMUST00000163827.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr10_-_59277570 | 0.37 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr9_-_107512566 | 0.37 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr2_+_51962831 | 0.36 |
ENSMUST00000112693.10
|
Rif1
|
replication timing regulatory factor 1 |
| chr7_+_128289261 | 0.36 |
ENSMUST00000151237.5
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr9_-_39863342 | 0.36 |
ENSMUST00000216647.2
|
Olfr975
|
olfactory receptor 975 |
| chr6_-_97408367 | 0.36 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B |
| chr2_+_22959452 | 0.36 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr11_-_70895213 | 0.36 |
ENSMUST00000124464.2
ENSMUST00000108527.8 |
Dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr2_-_152857239 | 0.36 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr11_+_57409484 | 0.35 |
ENSMUST00000108849.8
ENSMUST00000020830.14 |
Mfap3
|
microfibrillar-associated protein 3 |
| chr7_+_29467971 | 0.34 |
ENSMUST00000032802.5
|
Zfp84
|
zinc finger protein 84 |
| chr3_-_79439181 | 0.33 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr16_-_57427179 | 0.33 |
ENSMUST00000114371.5
ENSMUST00000232413.2 |
Cmss1
|
cms small ribosomal subunit 1 |
| chrX_-_100463395 | 0.33 |
ENSMUST00000117901.8
ENSMUST00000120201.8 ENSMUST00000117637.8 ENSMUST00000134005.2 ENSMUST00000121520.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr13_+_24798591 | 0.33 |
ENSMUST00000091694.10
|
Ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr1_+_179788675 | 0.33 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr18_-_84969601 | 0.33 |
ENSMUST00000025547.4
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr19_+_34078333 | 0.32 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr9_-_96601574 | 0.32 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr17_+_43671314 | 0.32 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr2_+_127750978 | 0.31 |
ENSMUST00000110344.2
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr13_+_23755551 | 0.31 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr7_+_99934685 | 0.31 |
ENSMUST00000057023.12
|
P4ha3
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide III |
| chr10_+_87926932 | 0.31 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chrM_+_11735 | 0.31 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr6_+_145899155 | 0.30 |
ENSMUST00000111701.2
|
Sspn
|
sarcospan |
| chr15_-_36792649 | 0.30 |
ENSMUST00000126184.2
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr14_-_49482846 | 0.30 |
ENSMUST00000227113.2
ENSMUST00000130853.2 ENSMUST00000228936.2 ENSMUST00000022398.15 |
ccdc198
|
coiled-coil domain containing 198 |
| chr9_+_3335135 | 0.30 |
ENSMUST00000212294.2
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr1_-_65162267 | 0.29 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr6_+_136530970 | 0.28 |
ENSMUST00000189535.2
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr17_+_71326542 | 0.28 |
ENSMUST00000179759.3
|
Myom1
|
myomesin 1 |
| chr8_+_88925821 | 0.27 |
ENSMUST00000066748.10
ENSMUST00000119033.8 ENSMUST00000118952.8 |
Tent4b
|
terminal nucleotidyltransferase 4B |
| chr6_+_67768007 | 0.27 |
ENSMUST00000196006.5
ENSMUST00000103307.3 |
Igkv14-130
|
immunoglobulin kappa variable 14-130 |
| chr8_-_25438784 | 0.26 |
ENSMUST00000119720.8
ENSMUST00000121438.9 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr1_-_87936242 | 0.25 |
ENSMUST00000187758.7
ENSMUST00000040783.11 |
Usp40
|
ubiquitin specific peptidase 40 |
| chr2_+_130509530 | 0.25 |
ENSMUST00000103193.5
|
Itpa
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr9_+_38766356 | 0.24 |
ENSMUST00000104874.3
|
Olfr26
|
olfactory receptor 26 |
| chr14_-_101846459 | 0.24 |
ENSMUST00000161991.8
|
Tbc1d4
|
TBC1 domain family, member 4 |
| chr12_-_83534482 | 0.24 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr15_-_102259158 | 0.23 |
ENSMUST00000231061.2
ENSMUST00000041208.9 |
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr6_-_69020489 | 0.23 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr13_-_8908798 | 0.23 |
ENSMUST00000176329.8
|
Wdr37
|
WD repeat domain 37 |
| chr11_+_118334297 | 0.23 |
ENSMUST00000133558.3
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr9_+_105272507 | 0.23 |
ENSMUST00000035181.10
ENSMUST00000123807.8 |
Aste1
|
asteroid homolog 1 |
| chr1_-_155910546 | 0.23 |
ENSMUST00000169241.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr2_-_26270612 | 0.22 |
ENSMUST00000114115.9
ENSMUST00000035427.11 |
Snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr11_-_110142565 | 0.21 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr7_-_43906802 | 0.21 |
ENSMUST00000107945.8
ENSMUST00000118216.8 |
Acp4
|
acid phosphatase 4 |
| chr6_+_67873135 | 0.20 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr3_+_68776884 | 0.20 |
ENSMUST00000054551.3
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr5_+_100187844 | 0.20 |
ENSMUST00000169390.8
ENSMUST00000031268.8 |
Enoph1
|
enolase-phosphatase 1 |
| chr10_+_57521930 | 0.20 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr14_+_61835675 | 0.20 |
ENSMUST00000165015.9
|
Trim13
|
tripartite motif-containing 13 |
| chr16_-_85347305 | 0.20 |
ENSMUST00000175700.8
ENSMUST00000114174.3 |
Cyyr1
|
cysteine and tyrosine-rich protein 1 |
| chr2_+_22959223 | 0.20 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr7_+_101512922 | 0.20 |
ENSMUST00000209334.2
|
Anapc15
|
anaphase promoting complex C subunit 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.6 | 3.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.5 | 2.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.5 | 3.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.5 | 1.4 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.4 | 1.6 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.4 | 1.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.4 | 1.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.4 | 2.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.4 | 1.1 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.4 | 1.1 | GO:1904959 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.4 | 1.4 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.4 | 1.1 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.3 | 2.4 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.3 | 2.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.0 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.3 | 1.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 0.8 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.2 | 2.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 2.8 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.2 | 0.9 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 1.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 1.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.9 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 3.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 2.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 5.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.4 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.7 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.5 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.1 | 0.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 1.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 1.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.3 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.1 | 1.6 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.9 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 1.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 3.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.7 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 3.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.4 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.5 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 2.0 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 5.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0070563 | cell migration involved in endocardial cushion formation(GO:0003273) negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.0 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.5 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.2 | GO:0070535 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 1.4 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.8 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.7 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.8 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.2 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.4 | 1.2 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.4 | 5.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 1.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 1.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 1.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 3.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 3.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.7 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.0 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 3.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.6 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 3.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.9 | 2.8 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.5 | 1.6 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.4 | 1.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 2.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.3 | 1.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.3 | 3.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.3 | 2.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 1.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 1.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 1.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 0.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 1.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 0.5 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.6 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 1.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.9 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.1 | 0.4 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 3.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 4.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 3.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 3.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 4.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 3.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 2.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |