Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
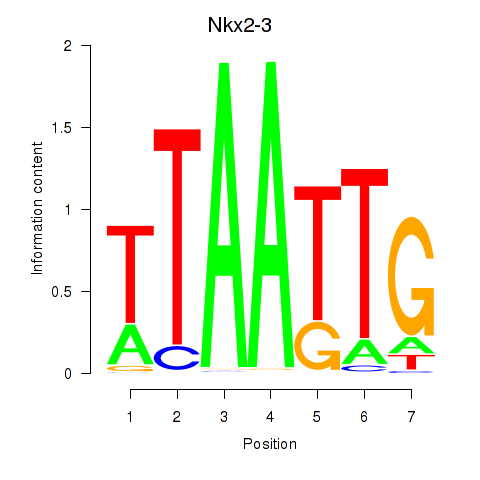
Results for Nkx2-3
Z-value: 1.26
Transcription factors associated with Nkx2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-3
|
ENSMUSG00000044220.14 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-3 | mm39_v1_chr19_+_43600738_43600764 | -0.71 | 1.4e-06 | Click! |
Activity profile of Nkx2-3 motif
Sorted Z-values of Nkx2-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_40078132 | 16.08 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr19_-_8382424 | 10.94 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr7_-_19432933 | 8.81 |
ENSMUST00000174355.8
ENSMUST00000172983.8 ENSMUST00000174710.2 ENSMUST00000003066.16 ENSMUST00000174064.9 |
Apoe
|
apolipoprotein E |
| chr5_-_87572060 | 8.80 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr10_+_87697155 | 7.85 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr2_+_70305267 | 7.46 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr19_+_37686240 | 7.05 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr7_-_48497771 | 6.72 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr3_-_137945419 | 6.69 |
ENSMUST00000199804.3
ENSMUST00000185122.8 ENSMUST00000183783.8 |
4930579F01Rik
0610031O16Rik
|
RIKEN cDNA 4930579F01 gene RIKEN cDNA 0610031O16 gene |
| chr19_-_39637489 | 5.43 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr14_-_118289557 | 4.88 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr19_-_4092218 | 4.75 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr5_+_146016064 | 4.66 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr6_+_125298372 | 4.62 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr5_+_42225303 | 4.12 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr17_+_37253802 | 4.08 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr6_+_125298296 | 3.87 |
ENSMUST00000081440.14
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr12_+_112073261 | 3.81 |
ENSMUST00000223412.2
|
Aspg
|
asparaginase |
| chr15_-_96929086 | 3.61 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr4_+_135870808 | 3.58 |
ENSMUST00000008016.3
|
Id3
|
inhibitor of DNA binding 3 |
| chr19_+_39980868 | 3.31 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr7_+_119217004 | 3.17 |
ENSMUST00000047929.13
ENSMUST00000135683.3 |
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr7_-_12731594 | 3.10 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr15_+_92495007 | 2.83 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr2_-_23939401 | 2.81 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr11_-_116080361 | 2.77 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr2_+_22959452 | 2.64 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr1_-_72323464 | 2.47 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chrM_+_5319 | 2.45 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr1_-_72323407 | 2.34 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr1_+_88034556 | 2.33 |
ENSMUST00000113137.2
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr15_+_31224460 | 2.29 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr15_-_50753792 | 2.20 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr1_+_13738967 | 2.15 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr6_+_54016543 | 2.09 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr6_+_141575226 | 2.08 |
ENSMUST00000042812.9
|
Slco1b2
|
solute carrier organic anion transporter family, member 1b2 |
| chr9_-_50657800 | 2.07 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr13_-_42001102 | 2.06 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr15_+_100768776 | 2.03 |
ENSMUST00000108909.9
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr8_+_46080840 | 2.02 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr18_+_20380397 | 2.00 |
ENSMUST00000054128.7
|
Dsg1c
|
desmoglein 1 gamma |
| chr16_-_23339548 | 1.97 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr10_-_12689345 | 1.91 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr15_-_77813123 | 1.90 |
ENSMUST00000109748.9
ENSMUST00000109747.9 ENSMUST00000100486.6 ENSMUST00000005487.12 |
Txn2
|
thioredoxin 2 |
| chr4_-_41640321 | 1.88 |
ENSMUST00000127306.2
|
Enho
|
energy homeostasis associated |
| chr17_+_56312672 | 1.84 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chrX_+_138464065 | 1.79 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr17_+_3447465 | 1.75 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr15_+_31224616 | 1.71 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr12_-_25147139 | 1.68 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr4_-_129155185 | 1.67 |
ENSMUST00000145261.8
|
C77080
|
expressed sequence C77080 |
| chr15_+_31225302 | 1.67 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr19_+_24853039 | 1.65 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr1_-_140111018 | 1.64 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr1_-_14374794 | 1.60 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_140111138 | 1.59 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr5_-_25305621 | 1.59 |
ENSMUST00000030784.14
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr2_+_22959223 | 1.58 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr2_-_63014622 | 1.58 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr10_+_29019645 | 1.55 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr8_-_5155347 | 1.55 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr14_+_67470735 | 1.55 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr6_-_83098255 | 1.55 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chr1_-_14374842 | 1.53 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr16_-_35589726 | 1.53 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr3_-_85648696 | 1.51 |
ENSMUST00000094148.6
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr6_+_125298168 | 1.50 |
ENSMUST00000176365.2
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr6_-_84565613 | 1.49 |
ENSMUST00000204146.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr11_+_99764215 | 1.49 |
ENSMUST00000093936.5
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr15_-_5093222 | 1.48 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr13_-_64277115 | 1.48 |
ENSMUST00000220792.2
ENSMUST00000222866.2 ENSMUST00000099441.6 ENSMUST00000222168.2 |
Slc35d2
|
solute carrier family 35, member D2 |
| chr3_+_55369149 | 1.48 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr2_+_125876883 | 1.45 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr2_-_32976378 | 1.44 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chrM_+_10167 | 1.44 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr2_+_69727563 | 1.43 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr15_+_31224555 | 1.42 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr10_-_85847697 | 1.41 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr13_+_23991010 | 1.40 |
ENSMUST00000006786.11
ENSMUST00000099697.3 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr11_+_94219046 | 1.38 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr5_-_28415020 | 1.37 |
ENSMUST00000118882.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr1_-_14380418 | 1.36 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_14380327 | 1.35 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr5_-_28415166 | 1.32 |
ENSMUST00000117098.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr19_+_31846154 | 1.31 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr19_-_10655391 | 1.31 |
ENSMUST00000025647.7
|
Pga5
|
pepsinogen 5, group I |
| chr8_-_64659004 | 1.27 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr11_+_94218810 | 1.26 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr12_-_101942463 | 1.25 |
ENSMUST00000221422.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr3_+_62327089 | 1.23 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr15_-_81244940 | 1.22 |
ENSMUST00000023040.9
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr15_+_58004831 | 1.22 |
ENSMUST00000226889.2
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr2_-_63014514 | 1.22 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr7_+_140181182 | 1.22 |
ENSMUST00000214180.2
ENSMUST00000211771.2 |
Olfr46
|
olfactory receptor 46 |
| chr19_+_44980565 | 1.22 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr2_+_80447389 | 1.22 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr15_+_100768806 | 1.21 |
ENSMUST00000201549.4
ENSMUST00000108908.6 |
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr13_-_43634695 | 1.20 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr2_-_147028309 | 1.20 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr3_+_60380243 | 1.19 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr7_-_35236962 | 1.17 |
ENSMUST00000193633.6
ENSMUST00000187190.7 ENSMUST00000205407.2 ENSMUST00000127472.3 |
Tdrd12
|
tudor domain containing 12 |
| chrX_+_141010919 | 1.14 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr2_-_113844100 | 1.14 |
ENSMUST00000090275.5
|
Gjd2
|
gap junction protein, delta 2 |
| chr6_-_148847633 | 1.13 |
ENSMUST00000132696.8
|
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr9_+_119231140 | 1.12 |
ENSMUST00000165044.3
|
Acvr2b
|
activin receptor IIB |
| chr1_+_66739990 | 1.11 |
ENSMUST00000027157.10
ENSMUST00000113995.2 |
Rpe
|
ribulose-5-phosphate-3-epimerase |
| chr17_-_78725510 | 1.11 |
ENSMUST00000234029.2
ENSMUST00000234530.2 ENSMUST00000234052.2 ENSMUST00000070039.14 ENSMUST00000112487.3 |
Fez2
|
fasciculation and elongation protein zeta 2 (zygin II) |
| chr2_+_152873772 | 1.08 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr8_+_120955195 | 1.07 |
ENSMUST00000180448.3
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr13_-_36918424 | 1.07 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
| chr19_+_60800012 | 1.06 |
ENSMUST00000128357.8
ENSMUST00000119633.8 ENSMUST00000025957.9 |
Dennd10
|
DENN domain containing 10 |
| chr9_-_44714263 | 1.06 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr15_+_102885467 | 1.06 |
ENSMUST00000001706.7
|
Hoxc9
|
homeobox C9 |
| chr18_-_39000056 | 1.05 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr11_-_59466995 | 1.05 |
ENSMUST00000215339.2
ENSMUST00000214351.2 |
Olfr222
|
olfactory receptor 222 |
| chr5_+_117919082 | 1.05 |
ENSMUST00000138579.3
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr18_+_37433852 | 1.04 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr1_+_66361252 | 1.04 |
ENSMUST00000123647.8
|
Map2
|
microtubule-associated protein 2 |
| chr1_-_63215952 | 1.04 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr4_+_135648041 | 1.03 |
ENSMUST00000030434.5
|
Fuca1
|
fucosidase, alpha-L- 1, tissue |
| chr6_-_115569504 | 1.02 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr6_-_136148820 | 1.00 |
ENSMUST00000188999.3
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr1_-_167294349 | 0.99 |
ENSMUST00000036643.6
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr9_+_46151994 | 0.99 |
ENSMUST00000034585.7
|
Apoa4
|
apolipoprotein A-IV |
| chr15_+_100768551 | 0.97 |
ENSMUST00000082209.13
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha |
| chr8_+_45960931 | 0.97 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr14_-_68771138 | 0.97 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr15_-_13173736 | 0.95 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr2_+_125876566 | 0.95 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr3_-_141687987 | 0.95 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr3_-_154036180 | 0.94 |
ENSMUST00000177846.8
|
Lhx8
|
LIM homeobox protein 8 |
| chr3_+_122213420 | 0.94 |
ENSMUST00000029766.9
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr2_+_57887896 | 0.94 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr6_-_41752111 | 0.93 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr7_-_12829100 | 0.92 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr5_+_81169049 | 0.92 |
ENSMUST00000117253.8
ENSMUST00000120128.8 |
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr3_+_85946145 | 0.91 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr13_-_53627110 | 0.91 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr15_+_58004753 | 0.91 |
ENSMUST00000067563.9
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr2_-_18053595 | 0.86 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr18_-_57108405 | 0.85 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr4_-_91264670 | 0.85 |
ENSMUST00000107109.9
ENSMUST00000107111.9 ENSMUST00000107120.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr15_+_58004793 | 0.84 |
ENSMUST00000227142.2
ENSMUST00000226955.2 |
Wdyhv1
|
WDYHV motif containing 1 |
| chrX_-_142716085 | 0.83 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr1_-_162687254 | 0.82 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_+_69727599 | 0.82 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr16_+_23338960 | 0.81 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr13_+_41013230 | 0.81 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr15_-_36165017 | 0.81 |
ENSMUST00000058643.4
|
Fbxo43
|
F-box protein 43 |
| chr16_+_8288627 | 0.80 |
ENSMUST00000046470.16
ENSMUST00000150790.2 ENSMUST00000142899.2 |
Mettl22
|
methyltransferase like 22 |
| chr15_-_36164963 | 0.80 |
ENSMUST00000227793.2
|
Fbxo43
|
F-box protein 43 |
| chr4_-_91264727 | 0.80 |
ENSMUST00000107124.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr1_-_162687369 | 0.79 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_+_83554770 | 0.78 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr14_+_75693396 | 0.78 |
ENSMUST00000164848.3
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr8_+_24159669 | 0.77 |
ENSMUST00000042352.11
ENSMUST00000207301.2 |
Zmat4
|
zinc finger, matrin type 4 |
| chr11_-_12362136 | 0.76 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr19_+_38253077 | 0.76 |
ENSMUST00000198045.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr19_+_38253105 | 0.76 |
ENSMUST00000196090.2
|
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr15_-_43733389 | 0.75 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr8_+_46081213 | 0.75 |
ENSMUST00000130850.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chrX_-_99638466 | 0.75 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr9_+_32027335 | 0.74 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_-_25200745 | 0.74 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_-_44541270 | 0.72 |
ENSMUST00000166808.2
|
Gm20538
|
predicted gene 20538 |
| chr6_-_138398376 | 0.72 |
ENSMUST00000163065.8
|
Lmo3
|
LIM domain only 3 |
| chr11_+_59197746 | 0.72 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr10_-_8632519 | 0.72 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr3_-_75177378 | 0.72 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr12_-_11486544 | 0.71 |
ENSMUST00000072299.7
|
Vsnl1
|
visinin-like 1 |
| chr3_+_55369384 | 0.71 |
ENSMUST00000200352.2
|
Dclk1
|
doublecortin-like kinase 1 |
| chr9_-_14411690 | 0.71 |
ENSMUST00000115647.3
|
Kdm4d
|
lysine (K)-specific demethylase 4D |
| chr2_+_67578556 | 0.70 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr7_+_144391786 | 0.70 |
ENSMUST00000155320.8
|
Fgf3
|
fibroblast growth factor 3 |
| chr11_-_99907030 | 0.70 |
ENSMUST00000018399.3
|
Krt33a
|
keratin 33A |
| chr1_-_119576347 | 0.69 |
ENSMUST00000027632.14
ENSMUST00000187194.2 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr1_+_19282278 | 0.69 |
ENSMUST00000064976.6
|
Tfap2b
|
transcription factor AP-2 beta |
| chr2_-_64806106 | 0.69 |
ENSMUST00000156765.2
|
Grb14
|
growth factor receptor bound protein 14 |
| chr18_-_84104507 | 0.69 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr1_+_183170293 | 0.69 |
ENSMUST00000192076.3
|
Taf1a
|
TATA-box binding protein associated factor, RNA polymerase I, A |
| chr2_+_22512195 | 0.68 |
ENSMUST00000028123.4
|
Gad2
|
glutamic acid decarboxylase 2 |
| chr12_+_116239006 | 0.68 |
ENSMUST00000090195.5
|
Gm11027
|
predicted gene 11027 |
| chr2_-_164013033 | 0.68 |
ENSMUST00000045196.4
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chrX_+_106193060 | 0.68 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr6_+_15185202 | 0.68 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr3_+_135053762 | 0.67 |
ENSMUST00000159658.8
ENSMUST00000078568.12 |
Slc9b1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr2_-_103114105 | 0.67 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chrM_+_7006 | 0.67 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr6_+_30512285 | 0.66 |
ENSMUST00000031798.14
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr5_+_8472831 | 0.66 |
ENSMUST00000066921.10
ENSMUST00000170496.6 |
Slc25a40
|
solute carrier family 25, member 40 |
| chr9_-_48747474 | 0.66 |
ENSMUST00000216150.2
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr15_-_50753437 | 0.66 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr13_-_104056803 | 0.66 |
ENSMUST00000091269.11
ENSMUST00000188997.7 ENSMUST00000169083.8 ENSMUST00000191275.7 |
Erbin
|
Erbb2 interacting protein |
| chr1_-_63215812 | 0.65 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr12_+_101370932 | 0.65 |
ENSMUST00000055156.5
|
Catsperb
|
cation channel sperm associated auxiliary subunit beta |
| chr2_-_86857424 | 0.65 |
ENSMUST00000214857.2
ENSMUST00000215972.2 |
Olfr1104
|
olfactory receptor 1104 |
| chr6_+_15184630 | 0.65 |
ENSMUST00000115470.3
|
Foxp2
|
forkhead box P2 |
| chr19_+_43770619 | 0.65 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr9_-_14411778 | 0.65 |
ENSMUST00000058796.7
|
Kdm4d
|
lysine (K)-specific demethylase 4D |
| chr5_+_73164226 | 0.64 |
ENSMUST00000031127.11
ENSMUST00000201304.2 |
Slc10a4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 2.2 | 6.7 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 1.6 | 4.8 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 1.5 | 4.5 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 1.3 | 16.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 1.3 | 3.8 | GO:0006530 | asparagine catabolic process(GO:0006530) |
| 1.2 | 8.5 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.1 | 7.9 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.8 | 2.5 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.8 | 3.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.7 | 10.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 6.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.6 | 2.8 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.6 | 2.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.6 | 1.7 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.5 | 1.6 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.5 | 2.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 1.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 1.3 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.4 | 1.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.4 | 4.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.4 | 3.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 1.0 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 0.3 | 1.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.3 | 8.0 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.3 | 0.9 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.3 | 8.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 1.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.3 | 1.9 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.3 | 1.3 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.3 | 1.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.5 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 0.7 | GO:0097275 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.2 | 0.7 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 1.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 10.3 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.2 | 0.6 | GO:0060300 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) |
| 0.2 | 1.2 | GO:0015867 | ATP transport(GO:0015867) |
| 0.2 | 1.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 2.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.6 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.2 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.3 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.2 | 0.6 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.2 | 0.8 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 2.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.5 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.2 | 1.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.6 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 | 4.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 4.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.7 | GO:0048319 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.5 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.7 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.3 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 1.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 6.8 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 0.7 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.5 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.2 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 1.9 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.5 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.5 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 1.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.5 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.0 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.4 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 1.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 1.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 1.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 4.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 1.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.4 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.8 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.1 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 1.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 2.8 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 1.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.6 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 1.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.7 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 1.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.4 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 4.7 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.1 | 1.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.4 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.1 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.1 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 4.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 1.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.9 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.3 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 2.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 1.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 1.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 1.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.7 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.8 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.0 | GO:0015881 | creatine transport(GO:0015881) |
| 0.0 | 0.6 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 2.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.7 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 2.8 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.0 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.0 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 2.0 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.3 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 2.1 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.1 | GO:0019236 | response to pheromone(GO:0019236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.8 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.0 | 4.8 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.8 | 7.9 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 14.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.3 | 1.3 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 0.6 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.2 | 1.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 1.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 3.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.3 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.1 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 4.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.9 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 8.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 2.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 3.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 3.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 1.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 3.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 6.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) outer dynein arm(GO:0036157) |
| 0.0 | 1.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.2 | GO:0035631 | IkappaB kinase complex(GO:0008385) CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 16.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.9 | 8.8 | GO:0046911 | metal chelating activity(GO:0046911) |
| 1.6 | 4.8 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 1.4 | 8.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.3 | 3.8 | GO:0004067 | asparaginase activity(GO:0004067) |
| 1.2 | 4.8 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.0 | 3.0 | GO:0070773 | protein-N-terminal glutamine amidohydrolase activity(GO:0070773) |
| 1.0 | 6.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.9 | 4.7 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.8 | 10.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.6 | 10.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 1.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.5 | 3.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.4 | 1.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.4 | 3.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.4 | 2.8 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.4 | 2.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.4 | 2.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 1.0 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.3 | 7.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 8.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 2.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 11.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.7 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.2 | 1.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 1.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 4.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 2.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 1.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 3.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 1.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 0.8 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.2 | 1.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 7.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 4.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 0.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 1.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 4.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 3.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 1.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 2.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 2.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.1 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 1.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.7 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 1.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 3.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 3.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 7.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 4.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 8.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 1.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 5.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 4.6 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 2.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 4.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 3.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 3.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |