Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
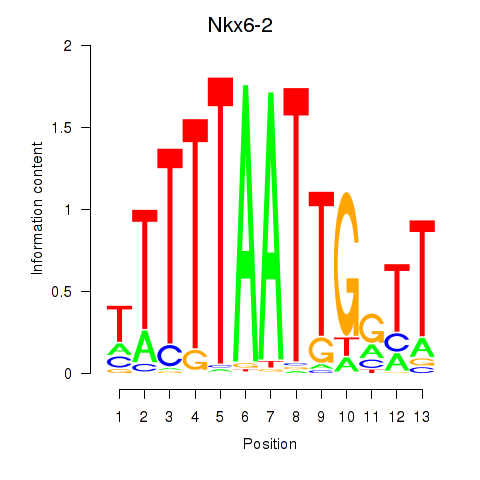
Results for Nkx6-2
Z-value: 1.25
Transcription factors associated with Nkx6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-2
|
ENSMUSG00000041309.18 | NK6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-2 | mm39_v1_chr7_-_139162706_139162724 | -0.26 | 1.2e-01 | Click! |
Activity profile of Nkx6-2 motif
Sorted Z-values of Nkx6-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142215027 | 16.17 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr7_-_142215595 | 12.39 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr14_+_80237691 | 9.81 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr19_+_58717319 | 9.00 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr3_-_10273628 | 8.92 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr10_+_115653152 | 7.93 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr5_-_110987604 | 7.20 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr5_+_110987839 | 7.17 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr9_+_96141317 | 6.84 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr9_+_96141299 | 6.78 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr9_+_96140781 | 6.54 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr18_-_74340885 | 6.28 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr4_-_117039809 | 6.09 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr4_+_140428777 | 5.97 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr18_+_34973605 | 5.83 |
ENSMUST00000043484.8
|
Reep2
|
receptor accessory protein 2 |
| chr7_+_89814713 | 5.73 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_+_66833434 | 5.69 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr16_-_21980200 | 5.41 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr12_+_117807607 | 5.17 |
ENSMUST00000176735.8
ENSMUST00000177339.2 |
Cdca7l
|
cell division cycle associated 7 like |
| chr10_-_37014859 | 5.13 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr18_-_74340842 | 5.12 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr5_-_122959321 | 4.95 |
ENSMUST00000197074.5
ENSMUST00000199406.5 ENSMUST00000196640.5 ENSMUST00000197719.5 ENSMUST00000200645.5 |
Anapc5
|
anaphase-promoting complex subunit 5 |
| chr5_-_110987441 | 4.85 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr17_+_36172210 | 4.46 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chrX_+_74425990 | 4.40 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr1_+_63216281 | 3.64 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr9_+_96140750 | 3.64 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr18_-_43610829 | 3.36 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr1_+_63215976 | 3.24 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr7_+_28488380 | 3.21 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr14_-_67246282 | 3.15 |
ENSMUST00000111115.8
ENSMUST00000022634.9 |
Bnip3l
|
BCL2/adenovirus E1B interacting protein 3-like |
| chr7_-_4400704 | 3.08 |
ENSMUST00000108590.4
ENSMUST00000206928.2 |
Gp6
|
glycoprotein 6 (platelet) |
| chr17_+_36172235 | 3.07 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr3_+_5815863 | 2.95 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr14_+_26722319 | 2.93 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr19_-_17350200 | 2.88 |
ENSMUST00000236139.2
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr8_-_65146079 | 2.80 |
ENSMUST00000048967.9
|
Cpe
|
carboxypeptidase E |
| chr2_-_174188505 | 2.80 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr11_+_20493306 | 2.67 |
ENSMUST00000093292.11
|
Sertad2
|
SERTA domain containing 2 |
| chr15_-_79718423 | 2.61 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr18_-_32044877 | 2.49 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr4_+_109200225 | 2.49 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr9_-_56151334 | 2.28 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr2_+_163535925 | 2.28 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr7_+_89780785 | 2.27 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_+_23206001 | 2.25 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr19_+_13208692 | 2.21 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr2_+_69691906 | 2.17 |
ENSMUST00000090852.11
ENSMUST00000166411.8 |
Ssb
|
Sjogren syndrome antigen B |
| chr14_+_51366512 | 2.00 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr9_-_123507847 | 1.96 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr1_-_126758369 | 1.90 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr17_-_78991691 | 1.89 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr15_-_80989200 | 1.86 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr5_+_75312939 | 1.78 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr9_-_22028370 | 1.75 |
ENSMUST00000213233.2
|
Elof1
|
ELF1 homolog, elongation factor 1 |
| chr8_-_62576140 | 1.71 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr7_-_15781838 | 1.67 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr4_+_3940747 | 1.65 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr2_+_3115250 | 1.65 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr17_-_30107544 | 1.54 |
ENSMUST00000171691.9
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr3_+_82962823 | 1.52 |
ENSMUST00000150268.8
ENSMUST00000122128.2 |
Plrg1
|
pleiotropic regulator 1 |
| chr5_-_21156766 | 1.48 |
ENSMUST00000036489.10
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr1_+_43484895 | 1.46 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr9_-_22028419 | 1.45 |
ENSMUST00000214394.2
ENSMUST00000013966.8 |
Elof1
|
ELF1 homolog, elongation factor 1 |
| chr1_-_149836974 | 1.44 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr10_+_79746690 | 1.41 |
ENSMUST00000181321.2
|
Gm26602
|
predicted gene, 26602 |
| chr6_-_87510200 | 1.39 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr11_-_73382303 | 1.38 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr19_-_14575395 | 1.35 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr2_+_170353338 | 1.34 |
ENSMUST00000136839.2
ENSMUST00000109148.8 ENSMUST00000170167.8 |
Pfdn4
|
prefoldin 4 |
| chr19_+_8779903 | 1.32 |
ENSMUST00000172175.3
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr8_+_21515561 | 1.31 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
| chr6_-_102441628 | 1.28 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr11_+_100750316 | 1.14 |
ENSMUST00000107356.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr3_+_96128427 | 1.14 |
ENSMUST00000090781.8
|
H2bc21
|
H2B clustered histone 21 |
| chr11_+_115824108 | 1.12 |
ENSMUST00000140991.2
|
Sap30bp
|
SAP30 binding protein |
| chr2_+_157209506 | 1.09 |
ENSMUST00000081202.6
|
Manbal
|
mannosidase, beta A, lysosomal-like |
| chrX_+_110801086 | 1.08 |
ENSMUST00000207962.2
|
Gm45194
|
predicted gene 45194 |
| chr2_+_85838122 | 1.06 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
| chr19_-_33764859 | 1.05 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr2_+_69050315 | 1.05 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr4_-_147726953 | 1.04 |
ENSMUST00000133006.2
ENSMUST00000037565.14 ENSMUST00000105720.8 |
Zfp979
|
zinc finger protein 979 |
| chr3_+_96508400 | 1.02 |
ENSMUST00000062058.5
|
Lix1l
|
Lix1-like |
| chr2_+_87609827 | 1.02 |
ENSMUST00000105210.3
|
Olfr152
|
olfactory receptor 152 |
| chr2_+_109522781 | 1.02 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr8_+_94537910 | 1.01 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr7_+_44711853 | 0.97 |
ENSMUST00000107829.9
ENSMUST00000003513.11 ENSMUST00000211465.2 ENSMUST00000210088.2 ENSMUST00000210520.2 |
Nosip
|
nitric oxide synthase interacting protein |
| chr5_+_123280250 | 0.97 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr17_-_48739874 | 0.96 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr14_+_27598021 | 0.94 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr11_-_109886601 | 0.92 |
ENSMUST00000020948.15
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr11_+_100750177 | 0.91 |
ENSMUST00000004145.14
ENSMUST00000133036.8 |
Stat5a
|
signal transducer and activator of transcription 5A |
| chr11_-_109886569 | 0.88 |
ENSMUST00000106669.3
|
Abca8b
|
ATP-binding cassette, sub-family A (ABC1), member 8b |
| chr11_+_99748741 | 0.88 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr2_-_125466985 | 0.88 |
ENSMUST00000089776.3
|
Cep152
|
centrosomal protein 152 |
| chr3_-_135373560 | 0.86 |
ENSMUST00000164430.7
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr8_+_21652293 | 0.85 |
ENSMUST00000098897.2
|
Defa22
|
defensin, alpha, 22 |
| chr13_+_83723743 | 0.84 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_+_85715984 | 0.83 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr15_+_34453432 | 0.76 |
ENSMUST00000060894.9
|
Erich5
|
glutamate rich 5 |
| chr11_-_107238956 | 0.74 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr10_+_26698556 | 0.69 |
ENSMUST00000135866.2
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr5_+_140404997 | 0.68 |
ENSMUST00000100507.8
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr4_-_133484080 | 0.65 |
ENSMUST00000008024.7
|
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr1_-_185061525 | 0.65 |
ENSMUST00000027921.11
ENSMUST00000110975.8 ENSMUST00000110974.4 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr19_-_12742811 | 0.64 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr7_+_107679062 | 0.63 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr9_-_113537277 | 0.61 |
ENSMUST00000111861.4
ENSMUST00000035086.13 |
Pdcd6ip
|
programmed cell death 6 interacting protein |
| chrX_-_42363663 | 0.61 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr1_-_64160557 | 0.60 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr8_-_49008305 | 0.59 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr4_+_146586445 | 0.59 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr13_+_110063364 | 0.57 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr3_+_106020545 | 0.57 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr5_+_25451771 | 0.56 |
ENSMUST00000144971.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr18_+_57275854 | 0.54 |
ENSMUST00000139892.2
|
Megf10
|
multiple EGF-like-domains 10 |
| chr2_-_37537224 | 0.54 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr7_-_84328553 | 0.54 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr4_-_154721288 | 0.53 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr11_-_17903861 | 0.50 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr18_+_4993795 | 0.49 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr4_+_147390131 | 0.49 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988 |
| chr2_-_87467879 | 0.48 |
ENSMUST00000216082.2
|
Olfr1132
|
olfactory receptor 1132 |
| chr5_+_43672856 | 0.48 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr1_-_126758520 | 0.48 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr7_-_19449319 | 0.45 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr2_-_88581690 | 0.44 |
ENSMUST00000215179.3
ENSMUST00000215529.3 |
Olfr1198
|
olfactory receptor 1198 |
| chr4_-_52859227 | 0.42 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273 |
| chr4_+_147637714 | 0.42 |
ENSMUST00000139784.8
ENSMUST00000143885.8 ENSMUST00000081742.7 |
Zfp985
|
zinc finger protein 985 |
| chr11_+_100902572 | 0.39 |
ENSMUST00000092663.4
|
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr9_-_21223631 | 0.39 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr19_-_13828056 | 0.39 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr8_+_22055402 | 0.39 |
ENSMUST00000084040.3
|
Defa37
|
defensin, alpha, 37 |
| chr11_-_115824290 | 0.36 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr3_+_69129745 | 0.35 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr7_+_4925781 | 0.34 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr9_+_51958453 | 0.34 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr13_+_83723255 | 0.33 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr8_+_21917427 | 0.33 |
ENSMUST00000095424.6
|
Defa36
|
defensin, alpha, 36 |
| chr1_-_171854818 | 0.30 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr2_-_88768449 | 0.29 |
ENSMUST00000215205.2
ENSMUST00000213412.2 |
Olfr1211
|
olfactory receptor 1211 |
| chr16_+_44215136 | 0.27 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr8_+_22155813 | 0.24 |
ENSMUST00000075268.5
|
Defa34
|
defensin, alpha, 34 |
| chr4_+_43851565 | 0.24 |
ENSMUST00000107860.3
|
Olfr155
|
olfactory receptor 155 |
| chr1_-_135513083 | 0.23 |
ENSMUST00000040599.15
|
Nav1
|
neuron navigator 1 |
| chr15_+_39255185 | 0.22 |
ENSMUST00000228839.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr16_-_16647139 | 0.22 |
ENSMUST00000023468.6
|
Spag6l
|
sperm associated antigen 6-like |
| chr2_-_131021905 | 0.22 |
ENSMUST00000089510.5
|
Cenpb
|
centromere protein B |
| chr19_-_13827773 | 0.21 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr5_-_131336914 | 0.19 |
ENSMUST00000160609.2
|
Galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr2_-_84480804 | 0.19 |
ENSMUST00000066177.10
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr4_-_147787010 | 0.19 |
ENSMUST00000117638.2
|
Zfp534
|
zinc finger protein 534 |
| chrX_+_132960055 | 0.19 |
ENSMUST00000144483.2
|
Cstf2
|
cleavage stimulation factor, 3' pre-RNA subunit 2 |
| chr5_+_20112704 | 0.18 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr10_+_129610507 | 0.17 |
ENSMUST00000203598.3
|
Olfr809
|
olfactory receptor 809 |
| chr4_+_148642879 | 0.16 |
ENSMUST00000017408.14
ENSMUST00000076022.7 |
Exosc10
|
exosome component 10 |
| chr14_-_31503869 | 0.16 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr4_+_147056433 | 0.14 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989 |
| chr2_+_24043159 | 0.13 |
ENSMUST00000028363.2
|
Il1f8
|
interleukin 1 family, member 8 |
| chr15_-_84804239 | 0.13 |
ENSMUST00000189185.2
|
Gm29666
|
predicted gene 29666 |
| chr6_-_81942906 | 0.13 |
ENSMUST00000032124.9
|
Mrpl19
|
mitochondrial ribosomal protein L19 |
| chr5_+_20112500 | 0.12 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_-_39188114 | 0.12 |
ENSMUST00000216698.2
|
Olfr945
|
olfactory receptor 945 |
| chr19_-_12313274 | 0.12 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chr4_+_145397238 | 0.12 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr7_-_102998876 | 0.10 |
ENSMUST00000215042.2
|
Olfr600
|
olfactory receptor 600 |
| chr4_+_147445744 | 0.09 |
ENSMUST00000133078.8
ENSMUST00000154154.2 |
Zfp978
|
zinc finger protein 978 |
| chr13_-_27697185 | 0.09 |
ENSMUST00000018389.5
ENSMUST00000110350.9 |
Prl8a8
|
prolactin family 8, subfamily a, member 81 |
| chr3_-_96359622 | 0.08 |
ENSMUST00000093126.11
ENSMUST00000098841.4 |
BC107364
|
cDNA sequence BC107364 |
| chrX_+_151922936 | 0.08 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chrX_+_99019176 | 0.05 |
ENSMUST00000113781.8
ENSMUST00000113783.8 ENSMUST00000113779.8 ENSMUST00000113776.8 ENSMUST00000113775.8 ENSMUST00000113780.8 ENSMUST00000113778.8 ENSMUST00000113777.8 ENSMUST00000071453.3 |
Eda
|
ectodysplasin-A |
| chr12_-_80807454 | 0.05 |
ENSMUST00000073251.8
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr8_+_21787455 | 0.03 |
ENSMUST00000098892.5
|
Defa5
|
defensin, alpha, 5 |
| chr3_+_68479578 | 0.02 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr2_-_89491811 | 0.02 |
ENSMUST00000215730.3
|
Olfr1250
|
olfactory receptor 1250 |
| chr4_+_151012375 | 0.01 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr11_+_98754434 | 0.01 |
ENSMUST00000142414.8
ENSMUST00000037480.9 |
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr2_+_85835884 | 0.01 |
ENSMUST00000111589.3
|
Olfr1032
|
olfactory receptor 1032 |
| chr14_+_51366306 | 0.00 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr15_+_65682066 | 0.00 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 28.6 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 2.2 | 6.7 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 2.0 | 6.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.5 | 12.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.3 | 8.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 1.1 | 6.7 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.0 | 6.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.0 | 5.8 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.9 | 2.8 | GO:0030070 | insulin processing(GO:0030070) |
| 0.5 | 2.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.5 | 3.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.5 | 2.1 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.5 | 2.0 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.5 | 8.9 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.4 | 1.8 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 1.5 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.3 | 0.9 | GO:1904632 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.3 | 2.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 1.4 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 2.5 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 2.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 5.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 0.6 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.2 | 0.6 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 1.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 9.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 3.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 3.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 7.9 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 5.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 6.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.7 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.0 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.6 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 2.9 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.6 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.5 | GO:0043654 | engulfment of apoptotic cell(GO:0043652) recognition of apoptotic cell(GO:0043654) |
| 0.1 | 9.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.6 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 2.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.3 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.5 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 4.4 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 1.0 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 1.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 2.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 23.2 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 0.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.2 | GO:0097151 | spontaneous neurotransmitter secretion(GO:0061669) positive regulation of inhibitory postsynaptic potential(GO:0097151) spontaneous synaptic transmission(GO:0098814) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 1.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.0 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 2.7 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 4.5 | GO:0006417 | regulation of translation(GO:0006417) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.0 | 6.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.9 | 5.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.8 | 11.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 9.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 7.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 5.9 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.3 | 3.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 4.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 2.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 2.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 5.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 4.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 6.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 23.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 1.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 5.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 1.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 6.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 1.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 2.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 5.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 6.6 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 2.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 3.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.6 | 29.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.6 | 1.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.6 | 2.9 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.5 | 8.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.5 | 9.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 1.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 6.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 1.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 12.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 6.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 3.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.2 | 0.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 2.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 5.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 2.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 2.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 3.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 6.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 4.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 4.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 5.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 3.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 26.8 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 3.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 4.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 8.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 10.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 5.0 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 2.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 2.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 5.7 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 2.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 6.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 5.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 2.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 3.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 25.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 6.7 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 23.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 6.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 5.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 8.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 2.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 2.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 28.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 6.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 5.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 8.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 5.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 12.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 25.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 7.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 5.4 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 7.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |