Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
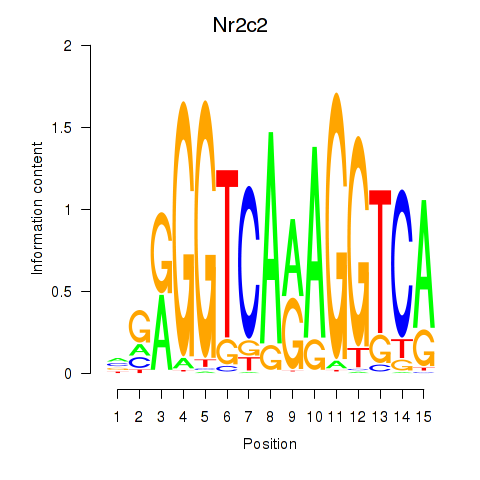
Results for Nr2c2
Z-value: 1.07
Transcription factors associated with Nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c2
|
ENSMUSG00000005893.15 | nuclear receptor subfamily 2, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c2 | mm39_v1_chr6_+_92068361_92068371 | -0.12 | 4.7e-01 | Click! |
Activity profile of Nr2c2 motif
Sorted Z-values of Nr2c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_9979033 | 4.73 |
ENSMUST00000121418.8
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr4_+_134591847 | 4.40 |
ENSMUST00000030627.8
|
Rhd
|
Rh blood group, D antigen |
| chr17_-_31348576 | 3.04 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr7_+_43086554 | 2.90 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr1_-_173161069 | 2.84 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr11_+_116089678 | 2.43 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr15_+_73620213 | 2.32 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr1_-_120047868 | 2.29 |
ENSMUST00000112648.8
|
Dbi
|
diazepam binding inhibitor |
| chr6_+_29694181 | 2.17 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr4_-_60697274 | 2.16 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr2_-_30720345 | 2.15 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr19_+_46120327 | 2.04 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr4_-_60457902 | 2.00 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr10_-_79624758 | 1.96 |
ENSMUST00000020573.13
|
Prss57
|
protease, serine 57 |
| chr16_+_33614378 | 1.95 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr4_-_62005498 | 1.94 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr4_-_133601990 | 1.92 |
ENSMUST00000168974.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr17_-_57137898 | 1.92 |
ENSMUST00000233000.2
ENSMUST00000002444.15 ENSMUST00000086801.7 |
Rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr16_+_33614715 | 1.79 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr10_+_75407356 | 1.77 |
ENSMUST00000143226.8
ENSMUST00000124259.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr16_+_48692976 | 1.76 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr7_+_43086432 | 1.74 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr11_+_61575245 | 1.71 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chr5_+_149215821 | 1.70 |
ENSMUST00000200928.2
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr5_+_114284585 | 1.70 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr3_-_89325594 | 1.68 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr4_-_61972348 | 1.66 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr11_+_58269862 | 1.66 |
ENSMUST00000013787.11
ENSMUST00000108826.3 |
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr12_-_79239022 | 1.62 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr11_+_120839879 | 1.56 |
ENSMUST00000154187.8
ENSMUST00000100130.10 ENSMUST00000129473.8 ENSMUST00000168579.8 |
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr15_-_102097387 | 1.46 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr11_+_4936824 | 1.45 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr2_+_118428690 | 1.44 |
ENSMUST00000038341.8
|
Bub1b
|
BUB1B, mitotic checkpoint serine/threonine kinase |
| chr5_-_120915693 | 1.43 |
ENSMUST00000044833.9
|
Oas3
|
2'-5' oligoadenylate synthetase 3 |
| chr6_-_83031358 | 1.40 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr10_-_81335966 | 1.39 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr9_-_57672375 | 1.39 |
ENSMUST00000215233.2
|
Clk3
|
CDC-like kinase 3 |
| chr2_+_24226857 | 1.39 |
ENSMUST00000114487.9
ENSMUST00000142093.7 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr10_-_93425553 | 1.38 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr7_+_126808016 | 1.37 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr3_+_51568588 | 1.35 |
ENSMUST00000099106.10
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr7_+_139673300 | 1.34 |
ENSMUST00000026540.9
|
Prap1
|
proline-rich acidic protein 1 |
| chr4_+_129355374 | 1.33 |
ENSMUST00000048162.10
ENSMUST00000138013.3 |
Bsdc1
|
BSD domain containing 1 |
| chr2_-_164198427 | 1.32 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr11_-_3454766 | 1.31 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr2_-_164197987 | 1.29 |
ENSMUST00000165980.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr9_-_22046970 | 1.29 |
ENSMUST00000165735.9
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr3_+_89043440 | 1.29 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr19_+_10019023 | 1.28 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr4_+_118286898 | 1.25 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr15_+_79230777 | 1.23 |
ENSMUST00000229130.2
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr6_+_124807176 | 1.22 |
ENSMUST00000131847.8
ENSMUST00000151674.3 |
Cdca3
|
cell division cycle associated 3 |
| chr6_-_83030759 | 1.22 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr7_-_126399574 | 1.22 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr3_+_89043879 | 1.21 |
ENSMUST00000107482.10
ENSMUST00000127058.2 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr9_+_110173253 | 1.21 |
ENSMUST00000199709.3
|
Scap
|
SREBF chaperone |
| chr16_-_4442802 | 1.21 |
ENSMUST00000014445.7
|
Pam16
|
presequence translocase-asssociated motor 16 homolog (S. cerevisiae) |
| chr7_-_44198157 | 1.21 |
ENSMUST00000145956.2
ENSMUST00000049343.15 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr7_-_132415257 | 1.20 |
ENSMUST00000097999.9
|
Fam53b
|
family with sequence similarity 53, member B |
| chr7_-_126399208 | 1.20 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr3_-_130524024 | 1.19 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr2_+_90912710 | 1.19 |
ENSMUST00000169852.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr2_+_174257597 | 1.18 |
ENSMUST00000109075.8
ENSMUST00000016397.7 |
Nelfcd
|
negative elongation factor complex member C/D, Th1l |
| chr10_-_62363217 | 1.18 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chr15_-_81756076 | 1.17 |
ENSMUST00000023117.10
|
Phf5a
|
PHD finger protein 5A |
| chr15_-_102097466 | 1.16 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr10_-_78080436 | 1.16 |
ENSMUST00000000384.8
|
Trappc10
|
trafficking protein particle complex 10 |
| chr3_+_51568625 | 1.15 |
ENSMUST00000159554.7
ENSMUST00000161590.4 |
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr8_-_106434565 | 1.15 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr2_+_163500290 | 1.13 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr7_-_48531344 | 1.10 |
ENSMUST00000119223.2
|
E2f8
|
E2F transcription factor 8 |
| chr4_-_116228921 | 1.09 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr17_+_47983587 | 1.09 |
ENSMUST00000152724.2
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr3_-_130523954 | 1.08 |
ENSMUST00000196202.5
ENSMUST00000133802.6 ENSMUST00000062601.14 ENSMUST00000200517.2 |
Rpl34
|
ribosomal protein L34 |
| chr11_-_69493567 | 1.07 |
ENSMUST00000138694.2
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr2_+_164647002 | 1.06 |
ENSMUST00000052107.5
|
Zswim3
|
zinc finger SWIM-type containing 3 |
| chr17_+_35268942 | 1.06 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr16_+_49675682 | 1.04 |
ENSMUST00000114496.3
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr8_+_71921824 | 1.04 |
ENSMUST00000124745.8
ENSMUST00000138892.2 ENSMUST00000147642.2 |
Dda1
|
DET1 and DDB1 associated 1 |
| chr1_+_63216281 | 1.03 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr19_-_4355983 | 1.02 |
ENSMUST00000025791.12
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr13_-_59917569 | 1.02 |
ENSMUST00000057115.7
|
Isca1
|
iron-sulfur cluster assembly 1 |
| chrX_-_72954835 | 1.00 |
ENSMUST00000114404.8
ENSMUST00000114407.9 ENSMUST00000114406.9 ENSMUST00000064376.13 ENSMUST00000114405.8 |
Arhgap4
|
Rho GTPase activating protein 4 |
| chr7_-_126399778 | 0.99 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr14_-_70414236 | 0.98 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr5_+_76988444 | 0.98 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr1_+_191553556 | 0.98 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr14_-_99231754 | 0.97 |
ENSMUST00000081987.5
|
Rpl36a-ps1
|
ribosomal protein L36A, pseudogene 1 |
| chr11_+_101333238 | 0.97 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chr1_-_120197979 | 0.96 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr1_+_63215976 | 0.93 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr3_+_137570248 | 0.93 |
ENSMUST00000041045.14
|
H2az1
|
H2A.Z variant histone 1 |
| chr5_+_31855009 | 0.92 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr10_+_81229443 | 0.92 |
ENSMUST00000118206.2
|
Smim24
|
small integral membrane protein 24 |
| chr1_+_175708341 | 0.91 |
ENSMUST00000195196.6
ENSMUST00000194306.6 ENSMUST00000193822.6 |
Exo1
|
exonuclease 1 |
| chr2_-_31973795 | 0.90 |
ENSMUST00000056406.7
|
Fam78a
|
family with sequence similarity 78, member A |
| chr2_-_93283024 | 0.90 |
ENSMUST00000111257.8
ENSMUST00000145553.8 |
Cd82
|
CD82 antigen |
| chr2_-_164646794 | 0.90 |
ENSMUST00000103094.11
ENSMUST00000017451.7 |
Acot8
|
acyl-CoA thioesterase 8 |
| chr1_-_136161850 | 0.90 |
ENSMUST00000120339.8
|
Inava
|
innate immunity activator |
| chr10_-_111838516 | 0.89 |
ENSMUST00000162508.9
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr1_+_136395673 | 0.89 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr3_+_107784543 | 0.89 |
ENSMUST00000037375.10
ENSMUST00000199990.2 |
Eps8l3
|
EPS8-like 3 |
| chr7_-_121620366 | 0.88 |
ENSMUST00000033160.15
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr6_+_85408953 | 0.88 |
ENSMUST00000045693.8
|
Smyd5
|
SET and MYND domain containing 5 |
| chr4_+_63477018 | 0.88 |
ENSMUST00000077709.11
|
Tmem268
|
transmembrane protein 268 |
| chr17_+_35069347 | 0.87 |
ENSMUST00000097343.11
ENSMUST00000173357.8 ENSMUST00000173065.8 ENSMUST00000165953.3 |
Nelfe
|
negative elongation factor complex member E, Rdbp |
| chr15_-_79967543 | 0.87 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr16_-_16687119 | 0.87 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr4_+_131600918 | 0.87 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr7_-_26895141 | 0.86 |
ENSMUST00000163311.9
ENSMUST00000126211.2 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr11_+_101333115 | 0.86 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr8_-_124045292 | 0.85 |
ENSMUST00000118395.2
ENSMUST00000035495.15 |
Fanca
|
Fanconi anemia, complementation group A |
| chr17_+_23945310 | 0.84 |
ENSMUST00000024701.9
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr5_-_109704339 | 0.83 |
ENSMUST00000198960.2
|
Crlf2
|
cytokine receptor-like factor 2 |
| chr5_+_31855394 | 0.80 |
ENSMUST00000063813.11
ENSMUST00000071531.12 ENSMUST00000131995.7 ENSMUST00000114507.10 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr1_+_87603952 | 0.80 |
ENSMUST00000170300.8
ENSMUST00000167032.2 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr14_+_75373766 | 0.80 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr15_-_100320926 | 0.80 |
ENSMUST00000023774.12
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr1_+_12788720 | 0.79 |
ENSMUST00000088585.10
|
Sulf1
|
sulfatase 1 |
| chr4_-_117146624 | 0.79 |
ENSMUST00000221654.2
|
Rnf220
|
ring finger protein 220 |
| chr4_+_133829898 | 0.78 |
ENSMUST00000070246.9
ENSMUST00000156750.2 |
Ubxn11
|
UBX domain protein 11 |
| chr15_+_81900570 | 0.77 |
ENSMUST00000069530.13
ENSMUST00000168581.8 ENSMUST00000164779.2 |
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr4_-_130068484 | 0.76 |
ENSMUST00000132545.3
ENSMUST00000175992.8 ENSMUST00000105999.9 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr7_-_19411866 | 0.76 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr11_+_51510555 | 0.76 |
ENSMUST00000127405.2
|
Nhp2
|
NHP2 ribonucleoprotein |
| chr5_+_92719336 | 0.76 |
ENSMUST00000176621.8
ENSMUST00000175974.2 ENSMUST00000131166.9 ENSMUST00000176448.8 ENSMUST00000082382.8 |
Fam47e
|
family with sequence similarity 47, member E |
| chr15_-_100976370 | 0.75 |
ENSMUST00000213610.2
|
Fignl2
|
fidgetin-like 2 |
| chr7_-_28297565 | 0.74 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr1_-_120198804 | 0.74 |
ENSMUST00000112641.8
|
Steap3
|
STEAP family member 3 |
| chr19_+_8907206 | 0.74 |
ENSMUST00000224272.2
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr5_+_31855304 | 0.72 |
ENSMUST00000114515.9
|
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr13_-_24945844 | 0.71 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chr17_-_25492340 | 0.71 |
ENSMUST00000173084.8
|
Ube2i
|
ubiquitin-conjugating enzyme E2I |
| chr4_+_118285275 | 0.70 |
ENSMUST00000006557.13
ENSMUST00000167636.8 ENSMUST00000102673.11 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr10_-_79820060 | 0.70 |
ENSMUST00000052885.14
|
Tmem259
|
transmembrane protein 259 |
| chr11_+_104498824 | 0.70 |
ENSMUST00000021028.5
|
Itgb3
|
integrin beta 3 |
| chr18_+_80250007 | 0.70 |
ENSMUST00000145963.9
ENSMUST00000025464.8 ENSMUST00000125127.8 ENSMUST00000025463.14 |
Txnl4a
Gm16286
|
thioredoxin-like 4A predicted gene 16286 |
| chr3_+_87989278 | 0.70 |
ENSMUST00000071812.11
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr6_-_30390996 | 0.70 |
ENSMUST00000152391.9
ENSMUST00000115184.2 ENSMUST00000080812.14 ENSMUST00000102992.10 |
Zc3hc1
|
zinc finger, C3HC type 1 |
| chr13_-_59823568 | 0.69 |
ENSMUST00000095739.10
ENSMUST00000022039.7 |
Golm1
|
golgi membrane protein 1 |
| chr7_-_143013899 | 0.69 |
ENSMUST00000208137.2
ENSMUST00000207910.2 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr2_+_180351910 | 0.69 |
ENSMUST00000029090.9
|
Gid8
|
GID complex subunit 8 |
| chr1_-_120048667 | 0.69 |
ENSMUST00000151708.3
|
Dbi
|
diazepam binding inhibitor |
| chr6_+_85428464 | 0.69 |
ENSMUST00000032078.9
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta) |
| chr17_+_35460722 | 0.68 |
ENSMUST00000068056.12
ENSMUST00000174757.8 ENSMUST00000173731.8 |
Ddx39b
|
DEAD box helicase 39b |
| chr13_-_76166789 | 0.68 |
ENSMUST00000179078.9
ENSMUST00000167271.9 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr4_-_120874376 | 0.68 |
ENSMUST00000043200.8
|
Smap2
|
small ArfGAP 2 |
| chr10_-_79819821 | 0.68 |
ENSMUST00000124536.2
|
Tmem259
|
transmembrane protein 259 |
| chr2_+_155453103 | 0.68 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr14_+_31881822 | 0.68 |
ENSMUST00000163336.8
ENSMUST00000169722.8 ENSMUST00000168385.8 |
Ncoa4
|
nuclear receptor coactivator 4 |
| chr11_-_54853729 | 0.68 |
ENSMUST00000108885.8
ENSMUST00000102730.9 ENSMUST00000018482.13 ENSMUST00000108886.8 ENSMUST00000102731.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr5_-_110928436 | 0.67 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr5_-_137305895 | 0.67 |
ENSMUST00000199243.5
ENSMUST00000197466.5 ENSMUST00000040873.12 |
Srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr5_-_113910571 | 0.67 |
ENSMUST00000019118.8
|
Sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr2_+_154498917 | 0.67 |
ENSMUST00000044277.10
|
Chmp4b
|
charged multivesicular body protein 4B |
| chr7_+_15832383 | 0.67 |
ENSMUST00000006181.7
|
Napa
|
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
| chr14_-_67953035 | 0.66 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr14_+_56091454 | 0.65 |
ENSMUST00000227465.2
ENSMUST00000168479.3 ENSMUST00000100529.10 |
Nynrin
|
NYN domain and retroviral integrase containing |
| chr8_+_70735477 | 0.65 |
ENSMUST00000087467.12
ENSMUST00000140212.8 ENSMUST00000110124.9 |
Homer3
|
homer scaffolding protein 3 |
| chr9_+_43957241 | 0.64 |
ENSMUST00000214627.2
|
Thy1
|
thymus cell antigen 1, theta |
| chr4_-_117539431 | 0.64 |
ENSMUST00000102687.4
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr13_-_73476561 | 0.64 |
ENSMUST00000222930.2
ENSMUST00000223293.2 ENSMUST00000022097.6 |
Ndufs6
|
NADH:ubiquinone oxidoreductase core subunit S6 |
| chr11_+_85202058 | 0.63 |
ENSMUST00000020835.16
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr8_+_79755194 | 0.63 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr17_-_48474356 | 0.62 |
ENSMUST00000027764.10
ENSMUST00000053612.14 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr4_-_3835595 | 0.62 |
ENSMUST00000138502.2
|
Rps20
|
ribosomal protein S20 |
| chr14_+_75373915 | 0.62 |
ENSMUST00000122840.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr6_+_136509922 | 0.62 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_-_8988854 | 0.62 |
ENSMUST00000042148.6
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr17_+_56610321 | 0.61 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr1_-_75101826 | 0.61 |
ENSMUST00000152855.3
|
Nhej1
|
non-homologous end joining factor 1 |
| chr19_-_4356207 | 0.61 |
ENSMUST00000088737.11
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr8_-_3671270 | 0.59 |
ENSMUST00000159548.2
ENSMUST00000019614.13 |
Xab2
|
XPA binding protein 2 |
| chr7_-_19684654 | 0.59 |
ENSMUST00000043440.8
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr2_-_157870341 | 0.59 |
ENSMUST00000029179.11
|
Tti1
|
TELO2 interacting protein 1 |
| chr10_+_81228877 | 0.59 |
ENSMUST00000105322.9
|
Smim24
|
small integral membrane protein 24 |
| chr8_-_94763634 | 0.59 |
ENSMUST00000212981.2
ENSMUST00000034204.11 |
Nudt21
|
nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
| chr9_+_20914211 | 0.59 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr17_-_25492508 | 0.59 |
ENSMUST00000174031.8
|
Ube2i
|
ubiquitin-conjugating enzyme E2I |
| chr18_+_21205386 | 0.58 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr18_-_74340885 | 0.58 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr3_-_89905927 | 0.58 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr17_-_35351026 | 0.58 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr10_-_87982732 | 0.58 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr17_+_26882171 | 0.58 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr3_+_96079642 | 0.56 |
ENSMUST00000076372.5
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr2_+_158636727 | 0.56 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr11_-_78313043 | 0.56 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr3_-_95041246 | 0.56 |
ENSMUST00000172572.9
ENSMUST00000173462.3 |
Scnm1
|
sodium channel modifier 1 |
| chr11_-_100986192 | 0.56 |
ENSMUST00000019447.15
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr8_+_96551974 | 0.56 |
ENSMUST00000074053.6
|
Sap18b
|
Sin3-associated polypeptide 18B |
| chr18_+_80249980 | 0.56 |
ENSMUST00000156400.9
|
Gm16286
|
predicted gene 16286 |
| chr3_+_14706781 | 0.56 |
ENSMUST00000029071.9
|
Car13
|
carbonic anhydrase 13 |
| chr18_+_80250102 | 0.55 |
ENSMUST00000127234.8
|
Gm16286
|
predicted gene 16286 |
| chr9_+_20914012 | 0.55 |
ENSMUST00000003386.7
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr19_-_61215743 | 0.55 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr16_-_17745999 | 0.55 |
ENSMUST00000003622.16
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr3_-_107992662 | 0.55 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr17_-_35978438 | 0.55 |
ENSMUST00000043674.15
|
Vars2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr8_-_84299850 | 0.55 |
ENSMUST00000212703.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.9 | 2.6 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.6 | 3.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.6 | 1.7 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.5 | 2.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.4 | 1.8 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.4 | 2.5 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.4 | 1.6 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 1.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.4 | 1.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 1.7 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.3 | 1.0 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.3 | 4.0 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 2.3 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.3 | 1.3 | GO:0033382 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.3 | 0.9 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 0.3 | 1.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 1.7 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.3 | 1.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 1.7 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.3 | 0.8 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.3 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.3 | 0.8 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.2 | 0.7 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 1.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 0.7 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.2 | 0.9 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 1.8 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 1.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 1.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 2.5 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 1.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 0.8 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 1.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.6 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.2 | 1.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 0.6 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.2 | 0.6 | GO:0034442 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.2 | 0.8 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.2 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 3.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.4 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.2 | 0.5 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.2 | 3.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 0.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.5 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.2 | 0.5 | GO:0006218 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.2 | 0.9 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 1.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 0.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 2.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.2 | 0.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 0.8 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.4 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.3 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.4 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.4 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 1.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 2.8 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:0014728 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.1 | 0.8 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 1.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 2.9 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 0.6 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.1 | 0.7 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.4 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.1 | 0.6 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.4 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 2.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.7 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 1.6 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.5 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.8 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.5 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.4 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 | 0.9 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.4 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 1.1 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 1.0 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 1.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.3 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 0.5 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.3 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.1 | 0.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 2.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.4 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 1.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.6 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.4 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.8 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.3 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.5 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0035854 | eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 0.9 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 0.1 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.1 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.4 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.1 | 1.0 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.9 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 1.9 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 2.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 0.3 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.2 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.7 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.1 | 1.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.4 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 3.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.2 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.1 | 0.7 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:2000556 | T-helper 1 cell cytokine production(GO:0035744) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.0 | 1.2 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.7 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.7 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.7 | GO:1904869 | regulation of establishment of protein localization to telomere(GO:0070203) protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.5 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 1.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 1.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 1.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 1.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.0 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 1.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.4 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 1.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 1.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.8 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.1 | GO:0032642 | regulation of chemokine production(GO:0032642) |
| 0.0 | 0.2 | GO:0097033 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 1.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 3.3 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.6 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:2000535 | exocyst assembly(GO:0001927) entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.0 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.0 | GO:0046144 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:1990879 | CST complex(GO:1990879) |
| 0.7 | 2.0 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.5 | 2.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.4 | 2.5 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.3 | 0.9 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.3 | 2.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 1.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 0.7 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.2 | 2.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 2.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 0.8 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 2.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.7 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) integrin alphav-beta3 complex(GO:0034683) |
| 0.2 | 1.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 2.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 3.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.9 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 1.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 3.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.7 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 1.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 5.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 2.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 2.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 3.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 3.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 7.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 2.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 6.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.7 | 3.7 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.6 | 2.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.5 | 1.6 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.5 | 2.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 1.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.4 | 1.3 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.4 | 2.5 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.4 | 2.6 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.3 | 3.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 1.7 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.3 | 1.0 | GO:0019978 | interleukin-3 receptor activity(GO:0004912) interleukin-3 binding(GO:0019978) |
| 0.3 | 4.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 2.3 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.3 | 1.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.3 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.2 | 0.7 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 1.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 1.7 | GO:0008823 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 1.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 0.8 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 1.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 1.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.7 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.5 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 1.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.8 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.8 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.5 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.4 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 4.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 2.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 3.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.9 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 7.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.2 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 1.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 2.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 6.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 2.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 6.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 4.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 5.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.8 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |