Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
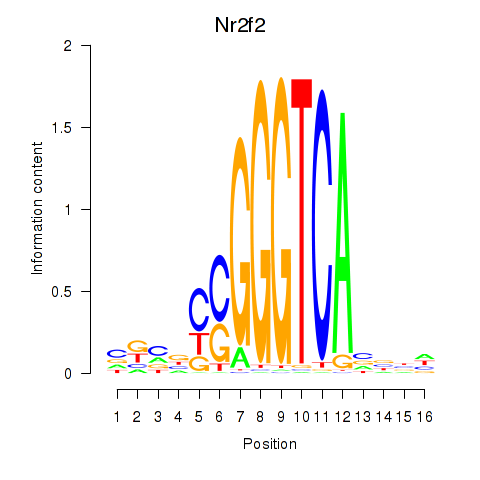
Results for Nr2f2
Z-value: 1.56
Transcription factors associated with Nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f2
|
ENSMUSG00000030551.15 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f2 | mm39_v1_chr7_-_70009669_70009697 | 0.29 | 8.7e-02 | Click! |
Activity profile of Nr2f2 motif
Sorted Z-values of Nr2f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_86577940 | 10.55 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr8_+_105996469 | 8.46 |
ENSMUST00000172525.8
ENSMUST00000174837.8 ENSMUST00000173859.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr17_-_56428968 | 8.25 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr7_-_30672747 | 7.86 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chrX_-_74918122 | 7.73 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr8_+_105996419 | 7.70 |
ENSMUST00000036127.9
ENSMUST00000163734.9 |
Hsf4
|
heat shock transcription factor 4 |
| chr7_-_30672824 | 7.52 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr7_-_30672889 | 7.37 |
ENSMUST00000001279.15
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr10_-_128509764 | 7.05 |
ENSMUST00000054764.9
|
Suox
|
sulfite oxidase |
| chr11_-_94492688 | 6.64 |
ENSMUST00000103164.4
|
Acsf2
|
acyl-CoA synthetase family member 2 |
| chr5_+_45650716 | 6.12 |
ENSMUST00000046122.11
|
Lap3
|
leucine aminopeptidase 3 |
| chr3_+_94600863 | 6.05 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr11_-_5865124 | 5.86 |
ENSMUST00000109823.9
ENSMUST00000109822.8 |
Gck
|
glucokinase |
| chr9_+_107454114 | 5.71 |
ENSMUST00000112387.9
ENSMUST00000123005.8 ENSMUST00000010195.14 ENSMUST00000144392.2 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr5_+_45650821 | 5.67 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr15_-_3612078 | 5.34 |
ENSMUST00000161770.2
|
Ghr
|
growth hormone receptor |
| chr5_+_135135735 | 5.18 |
ENSMUST00000201977.4
ENSMUST00000005507.10 |
Mlxipl
|
MLX interacting protein-like |
| chr17_-_57535003 | 5.05 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr1_-_91340884 | 4.56 |
ENSMUST00000086851.2
|
Hes6
|
hairy and enhancer of split 6 |
| chr16_-_10360893 | 4.43 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr2_+_68966125 | 4.27 |
ENSMUST00000041865.8
|
Nostrin
|
nitric oxide synthase trafficker |
| chrX_-_165262631 | 4.09 |
ENSMUST00000049435.15
|
Rab9
|
RAB9, member RAS oncogene family |
| chr1_-_183150867 | 3.88 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chr19_-_21630143 | 3.88 |
ENSMUST00000179768.8
ENSMUST00000178523.2 ENSMUST00000038830.10 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr19_-_59064501 | 3.82 |
ENSMUST00000163821.3
ENSMUST00000047511.15 |
Shtn1
|
shootin 1 |
| chr19_+_21630540 | 3.77 |
ENSMUST00000235332.2
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr8_+_60958931 | 3.58 |
ENSMUST00000079472.4
|
Aadat
|
aminoadipate aminotransferase |
| chr3_+_121517158 | 3.50 |
ENSMUST00000029771.13
|
F3
|
coagulation factor III |
| chr12_-_57592907 | 3.47 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr3_-_79536166 | 3.43 |
ENSMUST00000029386.14
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr19_+_31846154 | 3.42 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr11_-_70910058 | 3.41 |
ENSMUST00000108523.10
ENSMUST00000143850.8 |
Derl2
|
Der1-like domain family, member 2 |
| chr7_+_143028831 | 3.37 |
ENSMUST00000105917.3
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chrX_+_41157242 | 3.34 |
ENSMUST00000115095.9
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr5_+_65264863 | 3.33 |
ENSMUST00000204097.3
|
Klhl5
|
kelch-like 5 |
| chr2_-_179915276 | 3.31 |
ENSMUST00000108891.2
|
Cables2
|
CDK5 and Abl enzyme substrate 2 |
| chr12_-_103925197 | 3.31 |
ENSMUST00000122229.8
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr11_-_86884507 | 3.23 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr11_-_21521934 | 3.22 |
ENSMUST00000239073.2
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr1_+_181952302 | 3.18 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr6_-_119365632 | 3.16 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr1_+_9618173 | 3.15 |
ENSMUST00000144177.8
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr6_+_4600839 | 3.15 |
ENSMUST00000015333.12
ENSMUST00000181734.8 |
Casd1
|
CAS1 domain containing 1 |
| chr16_-_94657531 | 3.02 |
ENSMUST00000232562.2
ENSMUST00000165538.3 |
Kcnj6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr7_-_4633186 | 2.95 |
ENSMUST00000205360.2
ENSMUST00000206610.2 |
Tmem86b
|
transmembrane protein 86B |
| chr11_+_101358990 | 2.95 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr3_+_63202940 | 2.94 |
ENSMUST00000194150.6
|
Mme
|
membrane metallo endopeptidase |
| chr4_-_103072343 | 2.85 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr11_-_21522193 | 2.83 |
ENSMUST00000102874.11
ENSMUST00000238916.2 |
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr8_-_80021556 | 2.78 |
ENSMUST00000048718.4
|
Mmaa
|
methylmalonic aciduria (cobalamin deficiency) type A |
| chr10_-_128204545 | 2.77 |
ENSMUST00000220027.2
|
Coq10a
|
coenzyme Q10A |
| chr1_+_39026887 | 2.76 |
ENSMUST00000194552.2
|
Pdcl3
|
phosducin-like 3 |
| chr5_-_114582097 | 2.75 |
ENSMUST00000031560.14
|
Mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type homolog (human) |
| chr13_-_67081138 | 2.73 |
ENSMUST00000021991.11
|
Mterf3
|
mitochondrial transcription termination factor 3 |
| chr1_+_155911136 | 2.67 |
ENSMUST00000111757.10
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr11_+_97576724 | 2.58 |
ENSMUST00000107583.3
|
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr10_-_128204806 | 2.56 |
ENSMUST00000043211.7
ENSMUST00000220227.2 |
Coq10a
|
coenzyme Q10A |
| chr9_+_65536892 | 2.54 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr11_-_115188337 | 2.53 |
ENSMUST00000056153.8
|
Fads6
|
fatty acid desaturase domain family, member 6 |
| chr3_-_104127419 | 2.50 |
ENSMUST00000121198.8
ENSMUST00000122303.2 |
Magi3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr8_-_25506756 | 2.50 |
ENSMUST00000084032.6
ENSMUST00000207132.2 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr7_-_45109262 | 2.48 |
ENSMUST00000094434.13
|
Ftl1
|
ferritin light polypeptide 1 |
| chr5_+_121798932 | 2.45 |
ENSMUST00000111765.8
|
Brap
|
BRCA1 associated protein |
| chr15_-_76193955 | 2.40 |
ENSMUST00000210024.2
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr7_-_45109075 | 2.40 |
ENSMUST00000210864.2
|
Ftl1
|
ferritin light polypeptide 1 |
| chr8_-_25506916 | 2.40 |
ENSMUST00000084035.12
ENSMUST00000208247.3 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chrX_-_84820209 | 2.39 |
ENSMUST00000142152.2
ENSMUST00000156390.8 |
Gk
|
glycerol kinase |
| chr5_+_121798623 | 2.35 |
ENSMUST00000031414.15
|
Brap
|
BRCA1 associated protein |
| chr17_-_12726591 | 2.32 |
ENSMUST00000024595.4
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr5_-_114582053 | 2.32 |
ENSMUST00000123256.3
ENSMUST00000112245.6 |
Mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type homolog (human) |
| chr3_-_104127676 | 2.31 |
ENSMUST00000064371.14
|
Magi3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr19_+_21630887 | 2.30 |
ENSMUST00000052556.5
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr8_+_46428551 | 2.29 |
ENSMUST00000034051.7
ENSMUST00000150943.2 |
Ufsp2
|
UFM1-specific peptidase 2 |
| chr4_+_139350152 | 2.27 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr3_-_116505846 | 2.24 |
ENSMUST00000196335.2
ENSMUST00000120120.8 |
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr17_-_24915037 | 2.22 |
ENSMUST00000234235.2
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr17_+_25023263 | 2.22 |
ENSMUST00000234372.2
ENSMUST00000024972.7 |
Meiob
|
meiosis specific with OB domains |
| chr2_-_155849839 | 2.21 |
ENSMUST00000086145.10
ENSMUST00000144686.8 ENSMUST00000140657.8 |
6430550D23Rik
|
RIKEN cDNA 6430550D23 gene |
| chr19_-_6886965 | 2.20 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr9_+_107765320 | 2.17 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr1_+_155911518 | 2.16 |
ENSMUST00000133152.2
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr1_-_52766615 | 2.16 |
ENSMUST00000156876.8
ENSMUST00000087701.4 |
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr13_-_67080968 | 2.14 |
ENSMUST00000172597.8
ENSMUST00000173773.2 |
Mterf3
|
mitochondrial transcription termination factor 3 |
| chr2_+_126549239 | 2.14 |
ENSMUST00000028841.14
ENSMUST00000110416.3 |
Usp8
|
ubiquitin specific peptidase 8 |
| chrX_-_51254129 | 2.13 |
ENSMUST00000033450.3
|
Gpc4
|
glypican 4 |
| chr15_-_89080643 | 2.12 |
ENSMUST00000078953.9
|
Dennd6b
|
DENN/MADD domain containing 6B |
| chr7_+_4880111 | 2.10 |
ENSMUST00000125249.3
|
Isoc2a
|
isochorismatase domain containing 2a |
| chr19_-_6887361 | 2.10 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chr8_-_71112295 | 2.10 |
ENSMUST00000211715.2
ENSMUST00000210307.2 ENSMUST00000209768.2 ENSMUST00000070173.9 |
Pgpep1
|
pyroglutamyl-peptidase I |
| chr7_+_30673212 | 2.08 |
ENSMUST00000129773.2
|
Fam187b
|
family with sequence similarity 187, member B |
| chr8_+_85786684 | 2.08 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr6_-_120508250 | 2.06 |
ENSMUST00000075303.7
|
Hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr10_+_20024203 | 2.02 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr11_-_120687195 | 1.99 |
ENSMUST00000143139.8
ENSMUST00000129955.2 ENSMUST00000026151.11 ENSMUST00000167023.8 ENSMUST00000106133.8 ENSMUST00000106135.8 |
Dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr10_+_43777777 | 1.97 |
ENSMUST00000054418.12
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr5_+_137568086 | 1.97 |
ENSMUST00000198866.5
|
Tfr2
|
transferrin receptor 2 |
| chr5_-_108017019 | 1.96 |
ENSMUST00000128723.8
ENSMUST00000124034.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr5_+_137568113 | 1.96 |
ENSMUST00000031729.13
ENSMUST00000199054.5 |
Tfr2
|
transferrin receptor 2 |
| chr5_-_71815318 | 1.94 |
ENSMUST00000199357.2
|
Gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr16_+_20492014 | 1.94 |
ENSMUST00000154950.8
ENSMUST00000115461.8 ENSMUST00000136713.5 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr4_+_107035615 | 1.94 |
ENSMUST00000128123.3
ENSMUST00000106753.3 |
Tmem59
|
transmembrane protein 59 |
| chr17_+_6157154 | 1.93 |
ENSMUST00000149756.8
|
Tulp4
|
tubby like protein 4 |
| chr5_+_71815382 | 1.92 |
ENSMUST00000199967.5
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 |
| chr4_+_99952988 | 1.92 |
ENSMUST00000039630.6
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chrX_-_84820250 | 1.91 |
ENSMUST00000113978.9
|
Gk
|
glycerol kinase |
| chr3_-_79535966 | 1.89 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr2_+_31777273 | 1.89 |
ENSMUST00000138325.8
ENSMUST00000028187.7 |
Lamc3
|
laminin gamma 3 |
| chr19_+_11747721 | 1.87 |
ENSMUST00000167199.3
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr10_-_30494431 | 1.86 |
ENSMUST00000161074.8
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr15_+_9140614 | 1.85 |
ENSMUST00000227556.3
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chrX_+_41156713 | 1.84 |
ENSMUST00000115094.8
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr13_+_81034214 | 1.84 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr4_+_116414855 | 1.81 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr2_+_127112127 | 1.80 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr17_-_24915121 | 1.80 |
ENSMUST00000046839.10
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr13_+_67080864 | 1.79 |
ENSMUST00000021990.4
|
Ptdss1
|
phosphatidylserine synthase 1 |
| chr1_+_155911451 | 1.79 |
ENSMUST00000111754.9
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr8_-_47986473 | 1.76 |
ENSMUST00000039061.15
|
Trappc11
|
trafficking protein particle complex 11 |
| chr19_-_6886898 | 1.76 |
ENSMUST00000238095.2
|
Prdx5
|
peroxiredoxin 5 |
| chr13_+_93908138 | 1.76 |
ENSMUST00000091403.6
|
Arsb
|
arylsulfatase B |
| chr14_+_16508028 | 1.74 |
ENSMUST00000163885.2
|
Gm3248
|
predicted gene 3248 |
| chr17_+_15163446 | 1.74 |
ENSMUST00000052691.9
|
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene |
| chr19_+_44980565 | 1.73 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr7_-_18883113 | 1.73 |
ENSMUST00000032566.3
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr2_+_18681812 | 1.71 |
ENSMUST00000028071.13
|
Bmi1
|
Bmi1 polycomb ring finger oncogene |
| chr9_+_119168714 | 1.71 |
ENSMUST00000176351.8
|
Acaa1a
|
acetyl-Coenzyme A acyltransferase 1A |
| chr2_-_130266162 | 1.71 |
ENSMUST00000089581.11
|
Pced1a
|
PC-esterase domain containing 1A |
| chr7_-_127534601 | 1.70 |
ENSMUST00000141385.7
ENSMUST00000156152.3 |
Prss36
|
protease, serine 36 |
| chr6_-_149090146 | 1.70 |
ENSMUST00000095319.10
ENSMUST00000141346.2 ENSMUST00000111535.8 |
Amn1
|
antagonist of mitotic exit network 1 |
| chr16_-_11727262 | 1.70 |
ENSMUST00000127972.8
ENSMUST00000121750.2 ENSMUST00000096272.11 ENSMUST00000073371.7 |
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr19_-_21630078 | 1.70 |
ENSMUST00000177577.2
|
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr6_+_122285615 | 1.70 |
ENSMUST00000007602.15
ENSMUST00000112610.2 |
M6pr
|
mannose-6-phosphate receptor, cation dependent |
| chr3_+_63202687 | 1.70 |
ENSMUST00000194836.6
ENSMUST00000191633.6 |
Mme
|
membrane metallo endopeptidase |
| chrX_+_99818342 | 1.68 |
ENSMUST00000113735.3
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr19_-_42117420 | 1.68 |
ENSMUST00000161873.2
ENSMUST00000018965.4 |
Avpi1
|
arginine vasopressin-induced 1 |
| chrX_+_162691978 | 1.64 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr15_-_4008913 | 1.63 |
ENSMUST00000022791.9
|
Fbxo4
|
F-box protein 4 |
| chr6_+_115398996 | 1.62 |
ENSMUST00000000450.5
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr7_-_80052491 | 1.62 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr10_-_30494333 | 1.59 |
ENSMUST00000019925.7
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr2_-_165996716 | 1.58 |
ENSMUST00000139266.2
|
Sulf2
|
sulfatase 2 |
| chr1_-_13442658 | 1.58 |
ENSMUST00000081713.11
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr7_+_27878894 | 1.58 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr18_+_9212154 | 1.57 |
ENSMUST00000041080.7
|
Fzd8
|
frizzled class receptor 8 |
| chr5_+_21629932 | 1.57 |
ENSMUST00000056045.5
|
Fam185a
|
family with sequence similarity 185, member A |
| chr1_+_44187007 | 1.57 |
ENSMUST00000027214.4
|
Ercc5
|
excision repair cross-complementing rodent repair deficiency, complementation group 5 |
| chr6_-_24527545 | 1.57 |
ENSMUST00000118558.5
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr11_-_101875575 | 1.56 |
ENSMUST00000176261.2
ENSMUST00000143177.2 ENSMUST00000003612.13 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr4_+_24898074 | 1.56 |
ENSMUST00000029925.10
ENSMUST00000151249.2 |
Ndufaf4
|
NADH:ubiquinone oxidoreductase complex assembly factor 4 |
| chr9_-_44876817 | 1.55 |
ENSMUST00000214761.2
ENSMUST00000213666.2 ENSMUST00000213890.2 ENSMUST00000125642.8 ENSMUST00000213193.2 ENSMUST00000117506.9 ENSMUST00000138559.9 ENSMUST00000117549.8 |
Ube4a
|
ubiquitination factor E4A |
| chr3_+_154302311 | 1.55 |
ENSMUST00000192462.6
ENSMUST00000029850.15 |
Cryz
|
crystallin, zeta |
| chr18_+_11052458 | 1.55 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr9_-_105372235 | 1.54 |
ENSMUST00000176190.8
ENSMUST00000163879.9 ENSMUST00000112558.10 ENSMUST00000176363.9 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr17_+_6156738 | 1.54 |
ENSMUST00000142030.8
|
Tulp4
|
tubby like protein 4 |
| chr14_+_14901127 | 1.53 |
ENSMUST00000163790.2
|
Gm3558
|
predicted gene 3558 |
| chr6_+_4601124 | 1.53 |
ENSMUST00000141359.2
|
Casd1
|
CAS1 domain containing 1 |
| chr9_-_105372330 | 1.52 |
ENSMUST00000038118.15
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr6_+_59185846 | 1.49 |
ENSMUST00000062626.4
|
Tigd2
|
tigger transposable element derived 2 |
| chr17_+_15163466 | 1.49 |
ENSMUST00000164837.3
ENSMUST00000174004.2 |
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene |
| chr1_-_36748985 | 1.49 |
ENSMUST00000043951.10
|
Actr1b
|
ARP1 actin-related protein 1B, centractin beta |
| chr5_+_114582327 | 1.48 |
ENSMUST00000137167.8
ENSMUST00000112239.9 ENSMUST00000124260.8 ENSMUST00000125650.6 ENSMUST00000043760.15 |
Mvk
|
mevalonate kinase |
| chr12_-_101942172 | 1.48 |
ENSMUST00000221191.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr12_-_81579614 | 1.48 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr4_+_118266582 | 1.47 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr16_+_84631956 | 1.47 |
ENSMUST00000009120.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr16_-_17709457 | 1.47 |
ENSMUST00000117082.2
ENSMUST00000066127.13 ENSMUST00000117945.8 ENSMUST00000012152.13 |
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr14_-_19420488 | 1.47 |
ENSMUST00000166494.2
|
Gm2897
|
predicted gene 2897 |
| chr17_-_26463177 | 1.46 |
ENSMUST00000234687.2
ENSMUST00000234554.2 ENSMUST00000114988.8 ENSMUST00000141240.3 ENSMUST00000234175.2 |
Fam234a
|
family with sequence similarity 234, member A |
| chr8_+_77626400 | 1.45 |
ENSMUST00000109913.9
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr13_+_31809774 | 1.44 |
ENSMUST00000042054.3
|
Foxf2
|
forkhead box F2 |
| chr17_-_26463092 | 1.44 |
ENSMUST00000118487.8
ENSMUST00000234262.2 |
Fam234a
|
family with sequence similarity 234, member A |
| chr9_-_50605079 | 1.44 |
ENSMUST00000120622.2
|
Dixdc1
|
DIX domain containing 1 |
| chr15_+_9140685 | 1.44 |
ENSMUST00000090380.6
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr11_-_83189670 | 1.42 |
ENSMUST00000175741.8
ENSMUST00000176518.2 |
Pex12
|
peroxisomal biogenesis factor 12 |
| chr4_+_116415251 | 1.41 |
ENSMUST00000106475.2
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr14_+_16589391 | 1.40 |
ENSMUST00000164484.8
|
Gm8237
|
predicted gene 8237 |
| chr3_+_132389867 | 1.40 |
ENSMUST00000169172.5
|
Tbck
|
TBC1 domain containing kinase |
| chr7_-_19595221 | 1.38 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr1_-_185849448 | 1.38 |
ENSMUST00000045388.8
|
Lyplal1
|
lysophospholipase-like 1 |
| chr2_+_155224105 | 1.37 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr14_-_19261196 | 1.36 |
ENSMUST00000112797.12
ENSMUST00000225885.2 |
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr17_+_5891582 | 1.36 |
ENSMUST00000002436.11
|
Snx9
|
sorting nexin 9 |
| chr3_-_85653573 | 1.35 |
ENSMUST00000118408.8
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr17_+_8384333 | 1.35 |
ENSMUST00000097419.10
ENSMUST00000024636.15 |
Cep43
|
centrosomal protein 43 |
| chr17_-_24752683 | 1.35 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr8_+_53964721 | 1.35 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr2_-_26096547 | 1.34 |
ENSMUST00000028302.8
|
Lhx3
|
LIM homeobox protein 3 |
| chr4_-_148123223 | 1.33 |
ENSMUST00000030879.12
ENSMUST00000137724.8 |
Clcn6
|
chloride channel, voltage-sensitive 6 |
| chr19_+_56385531 | 1.33 |
ENSMUST00000026062.10
|
Casp7
|
caspase 7 |
| chr14_-_31552335 | 1.33 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr16_-_62667307 | 1.32 |
ENSMUST00000232561.2
ENSMUST00000089289.6 |
Arl13b
|
ADP-ribosylation factor-like 13B |
| chr10_-_43416941 | 1.32 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr7_+_27879650 | 1.31 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr2_+_24970327 | 1.31 |
ENSMUST00000044078.10
ENSMUST00000114380.9 |
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chrX_+_10581248 | 1.30 |
ENSMUST00000144356.8
|
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr12_-_101942463 | 1.30 |
ENSMUST00000221422.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr4_+_55350043 | 1.30 |
ENSMUST00000030134.9
|
Rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr11_-_83189734 | 1.29 |
ENSMUST00000108146.8
ENSMUST00000136369.2 ENSMUST00000018877.9 |
Pex12
|
peroxisomal biogenesis factor 12 |
| chr6_+_8259379 | 1.29 |
ENSMUST00000162034.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr6_+_8259405 | 1.28 |
ENSMUST00000160705.8
ENSMUST00000159433.8 |
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
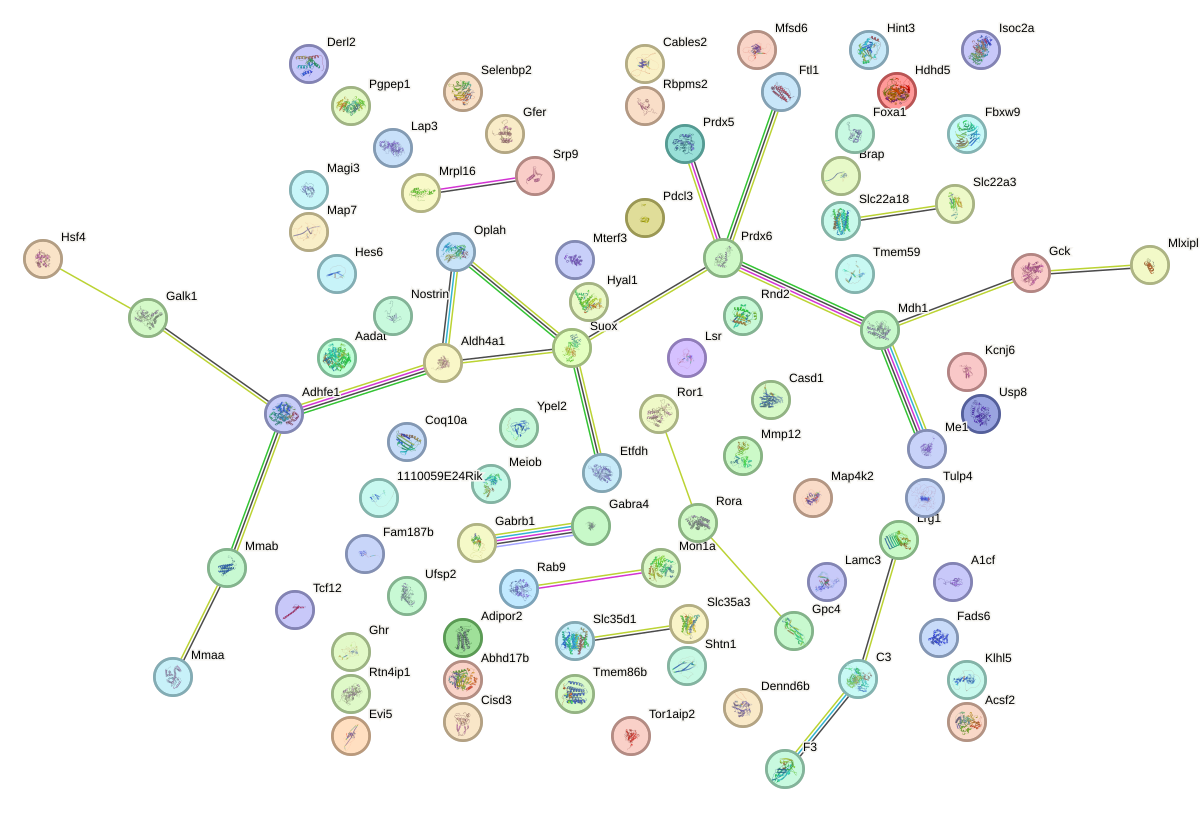
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 2.4 | 7.1 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 1.9 | 5.7 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.8 | 16.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 1.5 | 6.1 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 1.4 | 4.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 1.4 | 5.6 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 1.3 | 5.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.2 | 4.9 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 1.2 | 5.9 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 1.2 | 3.5 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.2 | 3.5 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.0 | 3.9 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 1.0 | 4.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.0 | 3.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.9 | 3.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.8 | 5.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.8 | 3.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.8 | 2.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.7 | 5.2 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.7 | 4.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.7 | 10.0 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.7 | 3.5 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.7 | 16.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.6 | 1.9 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.6 | 1.8 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.6 | 4.1 | GO:0052205 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.6 | 1.7 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.6 | 1.7 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.5 | 1.6 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.5 | 1.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.5 | 1.6 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.5 | 2.8 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.5 | 3.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.4 | 1.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.4 | 1.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.4 | 5.4 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.4 | 0.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 | 1.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 2.2 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.4 | 1.5 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.4 | 1.1 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.4 | 2.9 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.4 | 3.9 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 6.6 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 1.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.3 | 1.4 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.3 | 1.0 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.3 | 3.6 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.3 | 1.9 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.3 | 0.9 | GO:0043133 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) positive regulation of hindgut contraction(GO:0060450) regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 0.3 | 3.4 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 0.9 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 3.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.3 | 7.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 0.8 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.3 | 0.8 | GO:0022012 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.3 | 2.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.3 | 1.1 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.3 | 0.6 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.3 | 1.3 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 1.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 0.8 | GO:0061723 | glycophagy(GO:0061723) |
| 0.2 | 7.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 2.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 6.5 | GO:0006743 | ubiquinone metabolic process(GO:0006743) |
| 0.2 | 0.7 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 | 4.3 | GO:0051608 | histamine transport(GO:0051608) |
| 0.2 | 0.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 1.4 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.9 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 3.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.9 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 1.3 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.2 | 3.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.9 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.2 | 2.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 1.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.2 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 1.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 | 0.8 | GO:0000239 | pachytene(GO:0000239) |
| 0.2 | 0.6 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.2 | 1.5 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 2.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 0.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 0.9 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 0.9 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.5 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.2 | 0.5 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.2 | 1.6 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 0.9 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 1.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 3.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 1.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.2 | 1.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 2.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 1.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.2 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 0.5 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.2 | 1.8 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.6 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 2.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.7 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 0.9 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.6 | GO:0021679 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.1 | 0.6 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.6 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.4 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 0.8 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.3 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 1.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 1.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.4 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.1 | 2.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.7 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.8 | GO:2000053 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.9 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.9 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 1.0 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.6 | GO:0072181 | mesonephric duct formation(GO:0072181) |
| 0.1 | 1.9 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.6 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.9 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 3.8 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.3 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 1.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.1 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 1.5 | GO:0032927 | cytoplasm organization(GO:0007028) positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 1.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 2.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 1.3 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.1 | 3.0 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 1.0 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.7 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 4.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.8 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.2 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.1 | 0.5 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 1.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.4 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 | 0.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 6.3 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 1.5 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 1.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 3.4 | GO:0015893 | drug transport(GO:0015893) |
| 0.1 | 1.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 1.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 2.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.5 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.3 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.4 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 3.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.3 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 1.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 1.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 2.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 4.9 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 3.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 1.1 | GO:0002251 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 | 1.7 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 2.2 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.6 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 2.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 2.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.6 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 1.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.0 | 0.7 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.2 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 1.6 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 2.8 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.0 | 1.4 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 1.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 1.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:1902167 | cerebrospinal fluid secretion(GO:0033326) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.6 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 1.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.2 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 1.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.6 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.0 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.5 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.4 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.2 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 5.0 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 22.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.8 | 5.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.3 | 5.3 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.9 | 6.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.7 | 2.7 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.5 | 2.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.4 | 3.4 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.4 | 1.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 1.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.3 | 1.0 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.3 | 5.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 4.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 3.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 2.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 1.6 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.2 | 1.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 6.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 2.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 0.8 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 3.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 4.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 1.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 4.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.5 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 2.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 4.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 1.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.5 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 4.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 8.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 6.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 13.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 4.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 3.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 3.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 9.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 16.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 7.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 6.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 9.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 3.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 5.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 6.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 5.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 1.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 1.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 24.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 6.0 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.8 | 5.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 1.5 | 10.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.5 | 6.1 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 1.4 | 4.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 1.4 | 5.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.4 | 7.1 | GO:0030151 | molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 1.3 | 3.9 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 1.2 | 6.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.0 | 3.9 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.9 | 22.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.8 | 4.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.8 | 3.9 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.6 | 3.2 | GO:0097003 | adiponectin binding(GO:0055100) adipokinetic hormone receptor activity(GO:0097003) |
| 0.6 | 5.9 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.6 | 1.7 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.5 | 3.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.5 | 3.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.5 | 1.5 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.5 | 1.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.5 | 3.3 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.5 | 1.8 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.4 | 1.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.4 | 7.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 3.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.4 | 2.3 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.4 | 1.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.4 | 3.0 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.4 | 1.1 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.3 | 2.8 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 3.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 2.9 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.3 | 2.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.3 | 0.9 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.3 | 1.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 0.8 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.3 | 2.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.3 | 1.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.9 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 1.6 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 1.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 2.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 1.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 2.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.2 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.2 | 3.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 3.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 0.6 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.2 | 12.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.2 | 2.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 3.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 0.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 1.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 9.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.4 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 5.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 6.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 6.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 3.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 3.4 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 1.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.4 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 3.0 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 1.6 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 2.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 1.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.0 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 2.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.0 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 8.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 4.6 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.1 | 2.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 20.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 3.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 2.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.4 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 0.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 10.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 2.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 2.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0001727 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 8.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 3.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 1.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 6.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 2.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 3.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 4.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 3.4 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 1.1 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 3.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 6.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 1.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 13.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 9.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 4.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 5.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 11.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 3.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 5.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 5.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 3.4 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.3 | 2.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 3.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 2.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 6.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 1.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 5.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 5.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 11.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 4.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 6.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 19.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 5.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 3.6 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 3.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.5 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 3.2 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 1.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 2.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 4.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.7 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 3.2 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |