Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
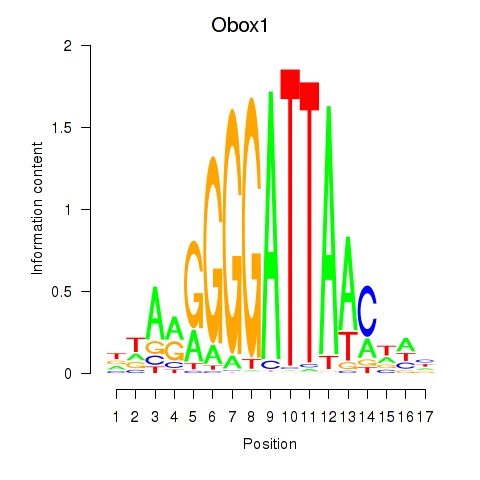
Results for Obox1
Z-value: 0.67
Transcription factors associated with Obox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Obox1
|
ENSMUSG00000054310.17 | oocyte specific homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Obox1 | mm39_v1_chr7_+_15280907_15280907 | -0.15 | 3.7e-01 | Click! |
Activity profile of Obox1 motif
Sorted Z-values of Obox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_46120327 | 6.27 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr6_-_41012435 | 6.00 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr4_-_137157824 | 5.57 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr19_-_8382424 | 5.48 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr8_+_114860375 | 5.23 |
ENSMUST00000147605.8
ENSMUST00000134593.2 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr15_+_76579885 | 4.99 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr15_+_76579960 | 4.94 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr8_+_110717062 | 4.73 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chr8_+_114860342 | 4.55 |
ENSMUST00000109109.8
|
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr6_+_30541581 | 4.37 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr11_+_3981769 | 4.22 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr19_-_40062174 | 4.03 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr1_-_139708906 | 4.03 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr9_-_48516447 | 3.85 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr13_+_55862437 | 3.45 |
ENSMUST00000021959.11
|
Txndc15
|
thioredoxin domain containing 15 |
| chr6_-_5193816 | 3.41 |
ENSMUST00000002663.12
|
Pon1
|
paraoxonase 1 |
| chr6_-_5193757 | 3.25 |
ENSMUST00000177159.9
ENSMUST00000176945.2 |
Pon1
|
paraoxonase 1 |
| chr16_-_17906886 | 3.24 |
ENSMUST00000132241.2
ENSMUST00000139861.2 ENSMUST00000003620.13 |
Prodh
|
proline dehydrogenase |
| chr14_-_31362835 | 3.13 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr1_-_139487951 | 3.11 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr4_-_137137088 | 2.86 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr19_-_8109346 | 2.76 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr9_-_86577940 | 2.62 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr14_+_66205932 | 2.40 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr6_+_41369290 | 2.34 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chr2_-_23939401 | 2.33 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr14_-_30665232 | 2.30 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr3_-_73615732 | 2.03 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr7_-_12732067 | 1.97 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr11_-_11848044 | 1.76 |
ENSMUST00000066237.10
|
Ddc
|
dopa decarboxylase |
| chr3_+_142406787 | 1.69 |
ENSMUST00000106218.8
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr11_-_94932158 | 1.68 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr3_-_63872189 | 1.59 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr3_+_142406827 | 1.57 |
ENSMUST00000044392.11
ENSMUST00000199519.5 |
Kyat3
|
kynurenine aminotransferase 3 |
| chr2_-_34990689 | 1.46 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr14_-_118289557 | 1.39 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr11_-_59937302 | 1.37 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr3_-_63872079 | 1.35 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr2_+_144435974 | 1.34 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr15_+_54975713 | 1.33 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr7_+_44033520 | 1.33 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr11_-_23615862 | 1.29 |
ENSMUST00000020523.4
|
Pex13
|
peroxisomal biogenesis factor 13 |
| chr19_+_7471398 | 1.27 |
ENSMUST00000170373.9
ENSMUST00000236308.2 ENSMUST00000235557.2 |
Atl3
|
atlastin GTPase 3 |
| chr7_-_67294943 | 1.23 |
ENSMUST00000190276.7
ENSMUST00000032775.12 ENSMUST00000053950.10 ENSMUST00000189836.2 |
Lrrc28
|
leucine rich repeat containing 28 |
| chr19_+_32573182 | 1.21 |
ENSMUST00000235594.2
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr11_-_62172164 | 1.18 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr13_-_64514830 | 1.13 |
ENSMUST00000222971.2
|
Ctsl
|
cathepsin L |
| chr17_+_79934096 | 1.09 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr15_-_76501041 | 1.08 |
ENSMUST00000073428.7
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr14_+_40826970 | 1.06 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr10_-_75616788 | 1.06 |
ENSMUST00000173512.2
ENSMUST00000173537.2 |
Gm20441
Gstt3
|
predicted gene 20441 glutathione S-transferase, theta 3 |
| chr1_+_128171859 | 0.99 |
ENSMUST00000027592.6
|
Ubxn4
|
UBX domain protein 4 |
| chr7_+_44034225 | 0.98 |
ENSMUST00000120262.2
|
Syt3
|
synaptotagmin III |
| chr15_-_76544308 | 0.98 |
ENSMUST00000066677.10
ENSMUST00000177359.2 |
Cyhr1
|
cysteine and histidine rich 1 |
| chrX_-_84820250 | 0.97 |
ENSMUST00000113978.9
|
Gk
|
glycerol kinase |
| chr7_+_81220987 | 0.95 |
ENSMUST00000165460.2
|
Whamm
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr11_-_101998648 | 0.94 |
ENSMUST00000177304.8
ENSMUST00000017455.15 |
Pyy
|
peptide YY |
| chr12_+_21366386 | 0.91 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chrX_+_141009756 | 0.89 |
ENSMUST00000112916.9
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr18_-_3281727 | 0.87 |
ENSMUST00000154705.8
ENSMUST00000151084.8 |
Crem
|
cAMP responsive element modulator |
| chr2_-_62476487 | 0.85 |
ENSMUST00000112459.4
ENSMUST00000028259.12 |
Ifih1
|
interferon induced with helicase C domain 1 |
| chr15_-_76501525 | 0.84 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr18_-_13013030 | 0.83 |
ENSMUST00000119512.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr15_+_89452529 | 0.82 |
ENSMUST00000023295.3
ENSMUST00000230538.2 ENSMUST00000230978.2 |
Acr
|
acrosin prepropeptide |
| chr10_+_61531282 | 0.81 |
ENSMUST00000020284.5
|
Tysnd1
|
trypsin domain containing 1 |
| chr18_+_67338437 | 0.79 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr7_+_142606476 | 0.79 |
ENSMUST00000037941.10
|
Cd81
|
CD81 antigen |
| chr19_+_44980565 | 0.78 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr19_+_34560922 | 0.75 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr9_+_57604895 | 0.74 |
ENSMUST00000034865.6
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr10_-_117582259 | 0.73 |
ENSMUST00000079041.7
|
Slc35e3
|
solute carrier family 35, member E3 |
| chr10_-_76949762 | 0.72 |
ENSMUST00000072755.12
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr18_-_3281752 | 0.72 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr6_-_71121324 | 0.71 |
ENSMUST00000074241.9
|
Thnsl2
|
threonine synthase-like 2 (bacterial) |
| chr2_+_152511381 | 0.71 |
ENSMUST00000125366.8
ENSMUST00000109825.8 ENSMUST00000089059.9 ENSMUST00000079247.4 |
H13
|
histocompatibility 13 |
| chr5_+_121798623 | 0.71 |
ENSMUST00000031414.15
|
Brap
|
BRCA1 associated protein |
| chr11_-_88742285 | 0.71 |
ENSMUST00000107903.8
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr9_-_106768601 | 0.67 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr7_-_7302468 | 0.67 |
ENSMUST00000000619.8
|
Clcn4
|
chloride channel, voltage-sensitive 4 |
| chr6_-_71121347 | 0.67 |
ENSMUST00000160918.8
|
Thnsl2
|
threonine synthase-like 2 (bacterial) |
| chr17_+_26780453 | 0.67 |
ENSMUST00000167662.8
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr19_+_24853039 | 0.65 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr11_-_53321242 | 0.63 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr7_+_16186704 | 0.63 |
ENSMUST00000019302.10
|
Tmem160
|
transmembrane protein 160 |
| chr6_-_122317156 | 0.62 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr8_+_91796680 | 0.62 |
ENSMUST00000034091.8
ENSMUST00000211136.2 |
Rbl2
|
RB transcriptional corepressor like 2 |
| chr1_-_136888118 | 0.61 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr9_+_123921573 | 0.60 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr2_-_102903680 | 0.59 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr3_+_87704258 | 0.58 |
ENSMUST00000029711.9
ENSMUST00000107582.3 |
Insrr
|
insulin receptor-related receptor |
| chr13_+_99481283 | 0.58 |
ENSMUST00000052249.7
ENSMUST00000224660.3 |
Mrps27
|
mitochondrial ribosomal protein S27 |
| chr1_+_140173787 | 0.57 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr4_-_140393185 | 0.57 |
ENSMUST00000069623.12
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr15_-_76544261 | 0.57 |
ENSMUST00000081291.13
|
Cyhr1
|
cysteine and histidine rich 1 |
| chr11_-_109613040 | 0.56 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr13_-_99481160 | 0.56 |
ENSMUST00000022153.8
|
Ptcd2
|
pentatricopeptide repeat domain 2 |
| chrX_+_141010919 | 0.56 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr14_-_30645711 | 0.54 |
ENSMUST00000006697.17
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr11_-_70350783 | 0.54 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr19_-_10079091 | 0.53 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr11_+_95275458 | 0.53 |
ENSMUST00000021243.16
ENSMUST00000146556.2 |
Slc35b1
|
solute carrier family 35, member B1 |
| chr17_+_44389704 | 0.53 |
ENSMUST00000154166.8
ENSMUST00000024756.5 |
Enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr2_+_19376447 | 0.52 |
ENSMUST00000023856.9
|
Msrb2
|
methionine sulfoxide reductase B2 |
| chr10_-_12744025 | 0.52 |
ENSMUST00000219660.2
|
Utrn
|
utrophin |
| chr9_-_35111172 | 0.52 |
ENSMUST00000176021.8
ENSMUST00000176531.8 ENSMUST00000176685.8 ENSMUST00000177129.8 |
Tirap
|
toll-interleukin 1 receptor (TIR) domain-containing adaptor protein |
| chr6_+_137387718 | 0.51 |
ENSMUST00000167002.4
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr7_-_18883113 | 0.51 |
ENSMUST00000032566.3
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr7_+_27879650 | 0.51 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr19_-_6919755 | 0.50 |
ENSMUST00000099782.10
|
Gpr137
|
G protein-coupled receptor 137 |
| chr7_+_37883216 | 0.50 |
ENSMUST00000177983.2
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr14_+_14090981 | 0.49 |
ENSMUST00000022269.7
|
Oit1
|
oncoprotein induced transcript 1 |
| chrX_-_8327965 | 0.49 |
ENSMUST00000103000.9
ENSMUST00000023931.4 |
Ssxb2
|
synovial sarcoma, X member B2 |
| chr7_+_43865369 | 0.49 |
ENSMUST00000074359.4
|
Klk1b5
|
kallikrein 1-related peptidase b5 |
| chr10_+_79977291 | 0.49 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr4_-_108158242 | 0.48 |
ENSMUST00000043616.7
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chr5_+_143450329 | 0.47 |
ENSMUST00000045593.12
|
Daglb
|
diacylglycerol lipase, beta |
| chr3_-_108053396 | 0.47 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr8_+_91796767 | 0.47 |
ENSMUST00000209518.2
|
Rbl2
|
RB transcriptional corepressor like 2 |
| chr6_+_48372771 | 0.47 |
ENSMUST00000114572.9
|
Krba1
|
KRAB-A domain containing 1 |
| chr7_+_37883300 | 0.47 |
ENSMUST00000179992.10
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene |
| chr4_+_155789246 | 0.47 |
ENSMUST00000030905.9
|
Ssu72
|
Ssu72 RNA polymerase II CTD phosphatase homolog (yeast) |
| chr2_+_155849965 | 0.46 |
ENSMUST00000006035.13
|
Ergic3
|
ERGIC and golgi 3 |
| chr14_+_14091030 | 0.46 |
ENSMUST00000224529.2
|
Oit1
|
oncoprotein induced transcript 1 |
| chr8_+_106434901 | 0.46 |
ENSMUST00000013302.7
ENSMUST00000211852.2 |
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr10_-_12743915 | 0.45 |
ENSMUST00000219584.2
|
Utrn
|
utrophin |
| chr7_-_134539993 | 0.44 |
ENSMUST00000171394.3
|
Insyn2a
|
inhibitory synaptic factor 2A |
| chr19_-_7183596 | 0.44 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr14_+_14210932 | 0.42 |
ENSMUST00000022271.14
|
Acox2
|
acyl-Coenzyme A oxidase 2, branched chain |
| chr2_-_180351778 | 0.42 |
ENSMUST00000103057.8
ENSMUST00000103055.8 |
Dido1
|
death inducer-obliterator 1 |
| chr6_-_59403264 | 0.42 |
ENSMUST00000051065.6
|
Gprin3
|
GPRIN family member 3 |
| chr8_-_104975134 | 0.42 |
ENSMUST00000212275.2
ENSMUST00000050211.7 |
Tk2
|
thymidine kinase 2, mitochondrial |
| chr15_+_76227695 | 0.42 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr17_+_56935118 | 0.41 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chr7_+_140500848 | 0.41 |
ENSMUST00000184560.2
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr17_+_37504783 | 0.41 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr9_-_64248570 | 0.41 |
ENSMUST00000068367.14
|
Dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr3_+_106628987 | 0.40 |
ENSMUST00000130105.2
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr6_+_70675416 | 0.40 |
ENSMUST00000103403.3
|
Igkv3-2
|
immunoglobulin kappa variable 3-2 |
| chr6_-_136150076 | 0.39 |
ENSMUST00000053880.13
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr11_-_103640640 | 0.39 |
ENSMUST00000018630.3
|
Wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr12_+_84820095 | 0.39 |
ENSMUST00000222022.2
|
Isca2
|
iron-sulfur cluster assembly 2 |
| chr4_-_34050076 | 0.39 |
ENSMUST00000029927.6
|
Spaca1
|
sperm acrosome associated 1 |
| chr16_+_33201164 | 0.38 |
ENSMUST00000165418.9
|
Zfp148
|
zinc finger protein 148 |
| chr19_-_7183626 | 0.37 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr6_+_124908389 | 0.37 |
ENSMUST00000180095.4
|
Mlf2
|
myeloid leukemia factor 2 |
| chr5_-_121798541 | 0.36 |
ENSMUST00000031412.12
ENSMUST00000111770.2 |
Acad10
|
acyl-Coenzyme A dehydrogenase family, member 10 |
| chrY_+_1010543 | 0.36 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr3_+_92123106 | 0.35 |
ENSMUST00000074449.7
ENSMUST00000090871.3 |
Sprr2a1
Sprr2a2
|
small proline-rich protein 2A1 small proline-rich protein 2A2 |
| chr14_-_30213408 | 0.35 |
ENSMUST00000112250.6
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr11_-_5657658 | 0.35 |
ENSMUST00000154330.2
|
Mrps24
|
mitochondrial ribosomal protein S24 |
| chr5_+_21942139 | 0.35 |
ENSMUST00000030882.12
|
Pmpcb
|
peptidase (mitochondrial processing) beta |
| chr17_+_33483650 | 0.34 |
ENSMUST00000217023.3
|
Olfr63
|
olfactory receptor 63 |
| chr7_-_80597479 | 0.34 |
ENSMUST00000026818.12
ENSMUST00000117383.8 ENSMUST00000119980.2 |
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr7_+_126396779 | 0.34 |
ENSMUST00000205324.2
|
Tlcd3b
|
TLC domain containing 3B |
| chr10_+_76284907 | 0.34 |
ENSMUST00000092406.12
|
2610028H24Rik
|
RIKEN cDNA 2610028H24 gene |
| chr7_-_80053063 | 0.34 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr7_+_44034028 | 0.34 |
ENSMUST00000130707.8
ENSMUST00000130844.3 |
Syt3
|
synaptotagmin III |
| chr7_+_81412695 | 0.33 |
ENSMUST00000133034.2
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr9_-_79667129 | 0.33 |
ENSMUST00000034881.8
|
Cox7a2
|
cytochrome c oxidase subunit 7A2 |
| chr7_-_28879879 | 0.33 |
ENSMUST00000209034.2
ENSMUST00000182328.8 ENSMUST00000186182.2 ENSMUST00000059642.17 |
Psmd8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr5_+_134212836 | 0.32 |
ENSMUST00000016086.10
|
Gtf2ird2
|
GTF2I repeat domain containing 2 |
| chr6_+_48372586 | 0.32 |
ENSMUST00000077093.7
|
Krba1
|
KRAB-A domain containing 1 |
| chr17_-_12726591 | 0.32 |
ENSMUST00000024595.4
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr10_+_79986988 | 0.32 |
ENSMUST00000146516.8
ENSMUST00000144526.2 |
Midn
|
midnolin |
| chr3_+_151143557 | 0.32 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr11_-_120687195 | 0.32 |
ENSMUST00000143139.8
ENSMUST00000129955.2 ENSMUST00000026151.11 ENSMUST00000167023.8 ENSMUST00000106133.8 ENSMUST00000106135.8 |
Dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr19_+_34585322 | 0.32 |
ENSMUST00000076249.6
|
Ifit3b
|
interferon-induced protein with tetratricopeptide repeats 3B |
| chr7_+_133311062 | 0.32 |
ENSMUST00000033282.5
|
Bccip
|
BRCA2 and CDKN1A interacting protein |
| chr6_+_124908341 | 0.32 |
ENSMUST00000203021.3
|
Mlf2
|
myeloid leukemia factor 2 |
| chr12_+_72132621 | 0.32 |
ENSMUST00000057257.10
ENSMUST00000117449.8 |
Jkamp
|
JNK1/MAPK8-associated membrane protein |
| chr4_+_56740070 | 0.31 |
ENSMUST00000181745.2
|
Gm26657
|
predicted gene, 26657 |
| chr2_-_62313981 | 0.30 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr13_+_58550499 | 0.29 |
ENSMUST00000225815.2
|
Rmi1
|
RecQ mediated genome instability 1 |
| chr12_+_84820024 | 0.29 |
ENSMUST00000021667.7
ENSMUST00000222449.2 ENSMUST00000222982.2 |
Isca2
|
iron-sulfur cluster assembly 2 |
| chr6_+_41155309 | 0.29 |
ENSMUST00000103276.3
|
Trbv19
|
T cell receptor beta, variable 19 |
| chr3_-_73615535 | 0.29 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
| chr14_-_77274056 | 0.28 |
ENSMUST00000062789.15
|
Lacc1
|
laccase domain containing 1 |
| chr17_-_35265702 | 0.28 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr14_-_55204383 | 0.28 |
ENSMUST00000111456.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr9_+_103917821 | 0.28 |
ENSMUST00000216593.2
ENSMUST00000147249.3 |
Nphp3
Gm28305
|
nephronophthisis 3 (adolescent) predicted gene 28305 |
| chr8_-_46428277 | 0.27 |
ENSMUST00000095323.8
ENSMUST00000098786.3 |
1700029J07Rik
|
RIKEN cDNA 1700029J07 gene |
| chr8_-_70975734 | 0.27 |
ENSMUST00000137610.3
|
Kxd1
|
KxDL motif containing 1 |
| chr7_+_7174315 | 0.27 |
ENSMUST00000051435.8
|
Zfp418
|
zinc finger protein 418 |
| chr17_-_25564501 | 0.27 |
ENSMUST00000153118.2
ENSMUST00000146856.3 |
Tpsab1
|
tryptase alpha/beta 1 |
| chr5_-_30401416 | 0.27 |
ENSMUST00000125367.4
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr1_+_178015287 | 0.26 |
ENSMUST00000159284.2
|
Desi2
|
desumoylating isopeptidase 2 |
| chr7_-_24287037 | 0.26 |
ENSMUST00000094705.3
|
Zfp575
|
zinc finger protein 575 |
| chr10_-_83369994 | 0.26 |
ENSMUST00000020497.14
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr11_+_96355475 | 0.26 |
ENSMUST00000107663.4
|
Skap1
|
src family associated phosphoprotein 1 |
| chr16_+_36514386 | 0.26 |
ENSMUST00000119464.2
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr2_+_83786037 | 0.25 |
ENSMUST00000179192.2
|
Gm13698
|
predicted gene 13698 |
| chr7_-_46445085 | 0.25 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr6_-_24515036 | 0.25 |
ENSMUST00000052277.5
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chr2_+_129642371 | 0.25 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr11_+_67857268 | 0.25 |
ENSMUST00000021286.11
ENSMUST00000108675.2 |
Stx8
|
syntaxin 8 |
| chr19_-_10859087 | 0.24 |
ENSMUST00000144681.2
|
Tmem109
|
transmembrane protein 109 |
| chr5_-_92822984 | 0.24 |
ENSMUST00000060930.10
|
Ccdc158
|
coiled-coil domain containing 158 |
| chr14_-_51033131 | 0.24 |
ENSMUST00000095932.5
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr4_-_155947819 | 0.24 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr14_-_34225281 | 0.24 |
ENSMUST00000171343.9
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.8 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 2.2 | 6.7 | GO:1902617 | response to fluoride(GO:1902617) |
| 1.1 | 3.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 4.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.5 | 8.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.5 | 4.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 6.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 2.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 2.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.5 | 1.4 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.4 | 1.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.4 | 2.4 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.4 | 2.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 1.9 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.4 | 1.1 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 0.3 | 3.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 4.0 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.3 | 2.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.3 | 1.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 1.2 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 1.4 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 1.8 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 0.7 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 0.7 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 | 3.3 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.2 | 1.4 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 0.6 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.2 | 0.7 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.2 | 0.5 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 0.5 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.2 | 0.5 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.8 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.0 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 2.6 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.6 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.8 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 1.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.4 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.1 | 1.1 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 0.3 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.3 | GO:0090472 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) dibasic protein processing(GO:0090472) |
| 0.1 | 0.2 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.4 | GO:0072181 | mesonephric duct formation(GO:0072181) |
| 0.1 | 0.6 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.6 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.8 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.1 | 0.3 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.1 | 2.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.8 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.2 | GO:0070537 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 8.4 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.2 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 | 0.5 | GO:0070940 | termination of RNA polymerase II transcription(GO:0006369) dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.4 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 3.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.8 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.3 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.5 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:2000864 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 2.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.2 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 1.0 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 1.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.5 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.0 | 0.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 1.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 1.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0071397 | regulation of response to osmotic stress(GO:0047484) cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.0 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.0 | 0.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.2 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.0 | 0.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.0 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.6 | 1.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 0.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 0.6 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 2.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.7 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 15.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 7.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 9.9 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 2.0 | 9.8 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.7 | 6.7 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 1.6 | 4.7 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.0 | 4.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.9 | 2.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.8 | 2.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.7 | 3.3 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.6 | 2.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.5 | 6.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 8.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.4 | 1.8 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.4 | 1.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 1.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 1.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 1.2 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 1.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 1.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 2.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.4 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.1 | 1.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 4.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 1.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.6 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.4 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 3.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 19.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.5 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.5 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.5 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.2 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.0 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 4.0 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 2.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 10.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 0.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 6.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 2.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 0.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 6.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |