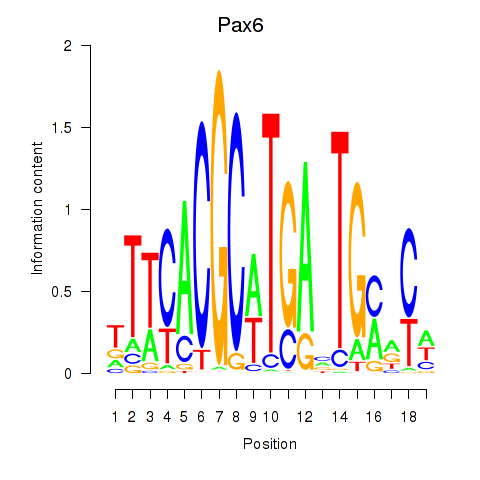
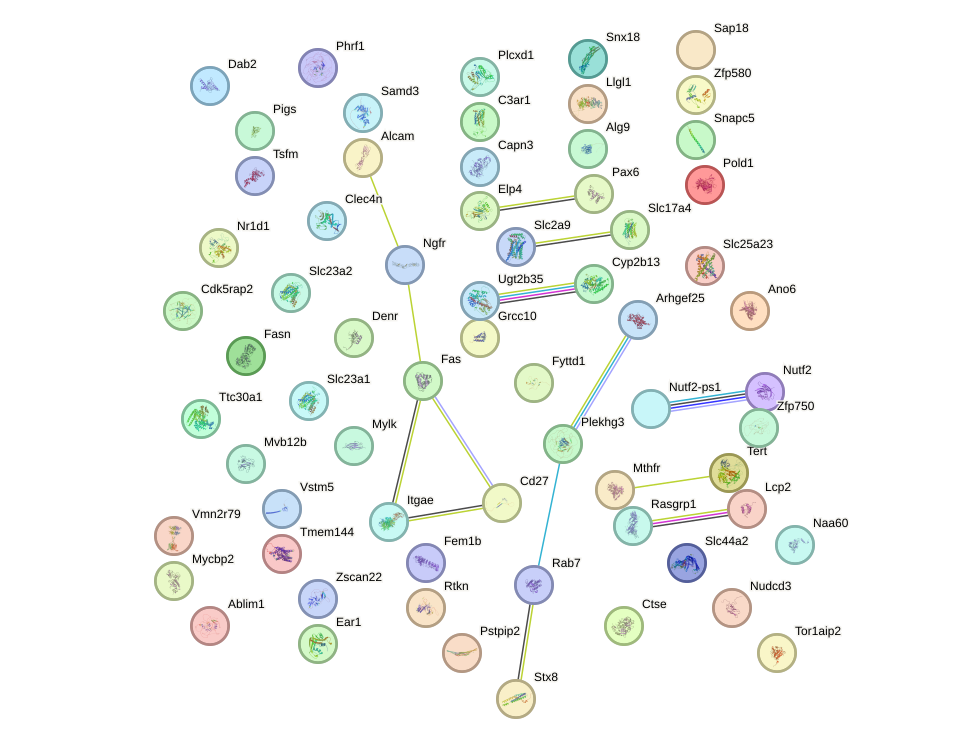
Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pax6
Z-value: 0.48
Transcription factors associated with Pax6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax6
|
ENSMUSG00000027168.22 | paired box 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax6 | mm39_v1_chr2_+_105499280_105499297 | 0.12 | 4.9e-01 | Click! |
Activity profile of Pax6 motif
Sorted Z-values of Pax6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_25760922 | 3.59 |
ENSMUST00000005669.9
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr14_-_44057096 | 1.85 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr13_-_24098951 | 1.70 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr13_-_24098981 | 1.66 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr13_-_4573312 | 1.48 |
ENSMUST00000221564.2
ENSMUST00000078239.5 ENSMUST00000080361.13 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr1_+_131566044 | 1.07 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr11_-_95478517 | 1.02 |
ENSMUST00000000122.7
|
Ngfr
|
nerve growth factor receptor (TNFR superfamily, member 16) |
| chr3_-_98660781 | 0.84 |
ENSMUST00000094050.11
ENSMUST00000090743.13 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr7_+_12631727 | 0.60 |
ENSMUST00000055528.11
ENSMUST00000117189.2 ENSMUST00000120809.2 ENSMUST00000119989.3 |
Zscan22
|
zinc finger and SCAN domain containing 22 |
| chr16_+_34605282 | 0.59 |
ENSMUST00000023538.9
|
Mylk
|
myosin, light polypeptide kinase |
| chr5_-_38637624 | 0.59 |
ENSMUST00000067886.12
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr17_+_25585255 | 0.56 |
ENSMUST00000234477.2
|
Tpsb2
|
tryptase beta 2 |
| chr15_+_6329263 | 0.52 |
ENSMUST00000078019.13
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr19_+_34268071 | 0.48 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr2_-_131987008 | 0.48 |
ENSMUST00000028815.15
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr4_-_70328659 | 0.47 |
ENSMUST00000144099.8
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chr5_-_38637474 | 0.45 |
ENSMUST00000143758.8
ENSMUST00000156272.8 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr15_+_6329278 | 0.44 |
ENSMUST00000159046.2
ENSMUST00000161040.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr19_+_34268053 | 0.43 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr11_+_33996920 | 0.42 |
ENSMUST00000052413.12
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr7_-_44198157 | 0.38 |
ENSMUST00000145956.2
ENSMUST00000049343.15 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr11_-_121410152 | 0.38 |
ENSMUST00000092298.6
|
Zfp750
|
zinc finger protein 750 |
| chr7_+_5054514 | 0.36 |
ENSMUST00000069324.7
|
Zfp580
|
zinc finger protein 580 |
| chr13_-_113755082 | 0.36 |
ENSMUST00000109241.5
|
Snx18
|
sorting nexin 18 |
| chr8_-_81607109 | 0.35 |
ENSMUST00000034150.10
|
Gab1
|
growth factor receptor bound protein 2-associated protein 1 |
| chr19_-_57107413 | 0.33 |
ENSMUST00000111528.8
ENSMUST00000111529.8 ENSMUST00000104902.9 |
Ablim1
|
actin-binding LIM protein 1 |
| chr16_-_52272828 | 0.33 |
ENSMUST00000170035.8
ENSMUST00000164728.8 ENSMUST00000168071.2 |
Alcam
|
activated leukocyte cell adhesion molecule |
| chr8_-_81607140 | 0.31 |
ENSMUST00000210676.2
|
Gab1
|
growth factor receptor bound protein 2-associated protein 1 |
| chr2_-_33777874 | 0.30 |
ENSMUST00000041555.10
|
Mvb12b
|
multivesicular body subunit 12B |
| chr6_-_122833109 | 0.28 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1 |
| chr4_+_148123554 | 0.26 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr11_+_72981377 | 0.24 |
ENSMUST00000006101.4
|
Itgae
|
integrin alpha E, epithelial-associated |
| chr5_+_87148697 | 0.24 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr6_+_123206802 | 0.24 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr13_+_73775001 | 0.24 |
ENSMUST00000022104.9
|
Tert
|
telomerase reverse transcriptase |
| chr15_+_95698574 | 0.22 |
ENSMUST00000226793.2
|
Ano6
|
anoctamin 6 |
| chr2_+_120331784 | 0.22 |
ENSMUST00000151342.3
|
Capn3
|
calpain 3 |
| chr7_+_140808680 | 0.22 |
ENSMUST00000106027.9
|
Phrf1
|
PHD and ring finger domains 1 |
| chr19_-_57107330 | 0.20 |
ENSMUST00000111526.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr4_+_155575792 | 0.20 |
ENSMUST00000165335.8
ENSMUST00000105616.10 ENSMUST00000030940.14 |
Gnb1
|
guanine nucleotide binding protein (G protein), beta 1 |
| chr10_-_127025851 | 0.19 |
ENSMUST00000222006.2
ENSMUST00000019611.15 ENSMUST00000219245.2 |
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr10_-_126866658 | 0.18 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr6_-_125213911 | 0.18 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chr9_+_21249118 | 0.18 |
ENSMUST00000034697.8
|
Slc44a2
|
solute carrier family 44, member 2 |
| chr2_-_75812311 | 0.18 |
ENSMUST00000099994.5
|
Ttc30a1
|
tetratricopeptide repeat domain 30A1 |
| chr9_-_62719208 | 0.17 |
ENSMUST00000034775.10
|
Fem1b
|
fem 1 homolog b |
| chr9_+_50686647 | 0.17 |
ENSMUST00000159576.2
|
Alg9
|
asparagine-linked glycosylation 9 (alpha 1,2 mannosyltransferase) |
| chr9_+_105520154 | 0.17 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr10_-_126866682 | 0.16 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr9_+_64086553 | 0.16 |
ENSMUST00000034965.8
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr9_+_15150341 | 0.15 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr2_-_117173190 | 0.14 |
ENSMUST00000173541.8
ENSMUST00000172901.8 ENSMUST00000173252.2 |
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr6_+_83119368 | 0.13 |
ENSMUST00000130622.8
ENSMUST00000129316.2 |
Rtkn
|
rhotekin |
| chr8_-_73229056 | 0.12 |
ENSMUST00000212991.2
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr5_+_110248276 | 0.11 |
ENSMUST00000141066.8
|
Plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr2_-_105734829 | 0.11 |
ENSMUST00000122965.8
|
Elp4
|
elongator acetyltransferase complex subunit 4 |
| chr13_+_63963054 | 0.10 |
ENSMUST00000021926.13
ENSMUST00000067821.13 ENSMUST00000144763.2 ENSMUST00000021925.14 ENSMUST00000238465.2 |
Ercc6l2
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 like 2 |
| chr8_-_73228953 | 0.09 |
ENSMUST00000079510.6
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr8_+_96551974 | 0.09 |
ENSMUST00000074053.6
|
Sap18b
|
Sin3-associated polypeptide 18B |
| chr3_-_79760067 | 0.09 |
ENSMUST00000192341.2
ENSMUST00000193410.3 |
Tmem144
|
transmembrane protein 144 |
| chr6_-_124718316 | 0.09 |
ENSMUST00000004389.6
|
Grcc10
|
gene rich cluster, C10 gene |
| chr2_-_117173312 | 0.09 |
ENSMUST00000178884.8
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr11_+_67857268 | 0.08 |
ENSMUST00000021286.11
ENSMUST00000108675.2 |
Stx8
|
syntaxin 8 |
| chr11_+_78219241 | 0.08 |
ENSMUST00000048073.9
|
Pigs
|
phosphatidylinositol glycan anchor biosynthesis, class S |
| chr17_-_57366795 | 0.08 |
ENSMUST00000040280.14
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr4_+_138920210 | 0.07 |
ENSMUST00000102508.10
ENSMUST00000131912.8 ENSMUST00000102507.10 |
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr11_+_104441489 | 0.07 |
ENSMUST00000018800.9
|
Myl4
|
myosin, light polypeptide 4 |
| chr7_+_75259778 | 0.07 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr5_-_134668152 | 0.07 |
ENSMUST00000036125.10
ENSMUST00000202622.4 |
Eif4h
|
eukaryotic translation initiation factor 4H |
| chr14_-_103583785 | 0.07 |
ENSMUST00000160758.8
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr11_-_6150411 | 0.07 |
ENSMUST00000066496.10
|
Nudcd3
|
NudC domain containing 3 |
| chr10_+_26105605 | 0.05 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr1_+_155916655 | 0.05 |
ENSMUST00000065648.15
ENSMUST00000097526.3 |
Tor1aip2
|
torsin A interacting protein 2 |
| chr18_+_77877611 | 0.05 |
ENSMUST00000238172.2
|
Pstpip2
|
proline-serine-threonine phosphatase-interacting protein 2 |
| chr6_-_88021999 | 0.04 |
ENSMUST00000113598.8
|
Rab7
|
RAB7, member RAS oncogene family |
| chr8_+_106587212 | 0.03 |
ENSMUST00000008594.9
|
Nutf2
|
nuclear transport factor 2 |
| chr5_+_124045238 | 0.03 |
ENSMUST00000023869.15
|
Denr
|
density-regulated protein |
| chr1_-_192812509 | 0.03 |
ENSMUST00000085555.7
|
Utp25
|
UTP25 small subunit processome component |
| chr16_+_3702523 | 0.02 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr12_+_76593799 | 0.02 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr11_+_60590584 | 0.02 |
ENSMUST00000108719.4
|
Llgl1
|
LLGL1 scribble cell polarity complex component |
| chr8_+_106587268 | 0.02 |
ENSMUST00000212610.2
ENSMUST00000212484.2 ENSMUST00000212200.2 |
Nutf2
|
nuclear transport factor 2 |
| chr7_+_86645323 | 0.01 |
ENSMUST00000233714.2
ENSMUST00000233648.2 ENSMUST00000164462.3 ENSMUST00000233730.2 |
Vmn2r79
|
vomeronasal 2, receptor 79 |
| chr5_+_124045552 | 0.01 |
ENSMUST00000166233.2
|
Denr
|
density-regulated protein |
| chr16_+_3702604 | 0.01 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr2_-_111104451 | 0.00 |
ENSMUST00000214760.2
|
Olfr1277
|
olfactory receptor 1277 |
| chr16_+_32698470 | 0.00 |
ENSMUST00000232272.2
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr11_+_60590498 | 0.00 |
ENSMUST00000052346.10
|
Llgl1
|
LLGL1 scribble cell polarity complex component |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.3 | 0.9 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.2 | 1.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 0.5 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 3.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.2 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 1.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.2 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.1 | 0.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 1.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.4 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.7 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 3.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.2 | GO:0045078 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.9 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.2 | 3.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 0.5 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.8 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 3.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 1.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 1.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |