Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Phox2a
Z-value: 0.20
Transcription factors associated with Phox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Phox2a
|
ENSMUSG00000007946.12 | paired-like homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Phox2a | mm39_v1_chr7_+_101467512_101467529 | 0.17 | 3.1e-01 | Click! |
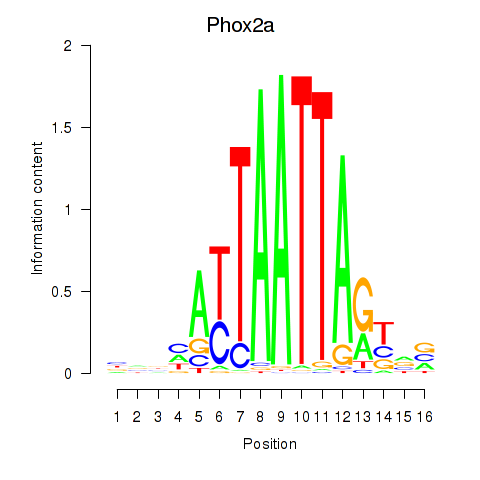
Activity profile of Phox2a motif
Sorted Z-values of Phox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7943365 | 0.95 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr6_+_37847721 | 0.30 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr11_-_99134885 | 0.18 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr11_+_116734104 | 0.16 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr9_+_118892497 | 0.16 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chrX_+_9751861 | 0.13 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr3_+_66127330 | 0.10 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr6_-_30936013 | 0.10 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr2_-_89855921 | 0.10 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr10_-_129965752 | 0.09 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr5_+_13448647 | 0.09 |
ENSMUST00000125629.8
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr10_+_29074950 | 0.09 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr19_-_55229668 | 0.08 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g |
| chr5_+_13448833 | 0.08 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr5_-_34817412 | 0.08 |
ENSMUST00000041364.13
|
Nop14
|
NOP14 nucleolar protein |
| chr4_+_138606671 | 0.08 |
ENSMUST00000105804.2
|
Pla2g2e
|
phospholipase A2, group IIE |
| chr2_+_36120438 | 0.08 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr3_-_72875187 | 0.07 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr11_-_99979052 | 0.07 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr1_-_154692678 | 0.07 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr19_+_12364643 | 0.07 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chr12_+_30934320 | 0.07 |
ENSMUST00000067087.7
|
Alkal2
|
ALK and LTK ligand 2 |
| chr16_-_29363671 | 0.06 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr13_-_78344492 | 0.06 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr4_-_43710231 | 0.06 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr13_-_22289994 | 0.06 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr14_+_26722319 | 0.06 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr2_-_111820618 | 0.06 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr5_-_118382926 | 0.05 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr17_-_37430949 | 0.05 |
ENSMUST00000214994.2
ENSMUST00000216341.2 |
Olfr92
|
olfactory receptor 92 |
| chr19_-_47680528 | 0.05 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr1_+_173983199 | 0.04 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr2_-_164427367 | 0.04 |
ENSMUST00000109342.2
|
Wfdc6a
|
WAP four-disulfide core domain 6A |
| chr7_-_44752508 | 0.04 |
ENSMUST00000209830.2
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr16_-_56688024 | 0.04 |
ENSMUST00000232373.2
|
Tmem45a
|
transmembrane protein 45a |
| chr4_+_134658209 | 0.04 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr3_+_93301003 | 0.04 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chr2_-_168607166 | 0.03 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr2_-_111843053 | 0.03 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr8_-_3675024 | 0.03 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr15_+_98350469 | 0.03 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr10_-_128885867 | 0.03 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr14_-_68771138 | 0.03 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr6_-_41752111 | 0.03 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr1_+_173924457 | 0.03 |
ENSMUST00000213832.2
|
Olfr427
|
olfactory receptor 427 |
| chr6_+_92793440 | 0.03 |
ENSMUST00000057977.4
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr19_-_45224251 | 0.03 |
ENSMUST00000099401.6
|
Lbx1
|
ladybird homeobox 1 |
| chr2_-_27365633 | 0.02 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr4_-_14621497 | 0.02 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr8_-_85389470 | 0.02 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2 |
| chr5_-_66672158 | 0.02 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr14_+_65612788 | 0.02 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chrX_+_159551171 | 0.01 |
ENSMUST00000112368.3
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr4_-_14621805 | 0.01 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr13_-_103042294 | 0.01 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr4_-_14621669 | 0.00 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr6_-_147165623 | 0.00 |
ENSMUST00000052296.9
ENSMUST00000204197.2 |
Pthlh
|
parathyroid hormone-like peptide |
| chrX_+_159551009 | 0.00 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Phox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.0 | 0.1 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |