Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
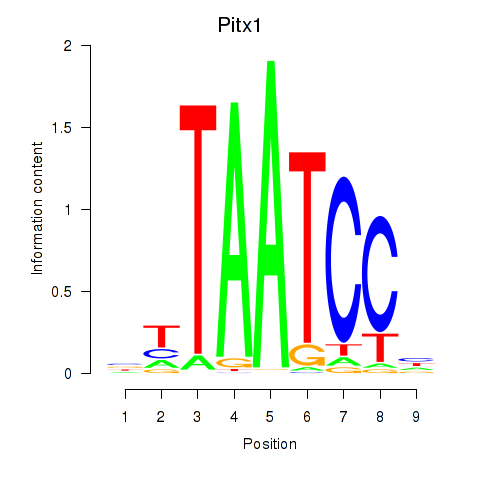
Results for Pitx1
Z-value: 1.05
Transcription factors associated with Pitx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx1
|
ENSMUSG00000021506.8 | paired-like homeodomain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx1 | mm39_v1_chr13_-_55983946_55984005 | -0.14 | 4.2e-01 | Click! |
Activity profile of Pitx1 motif
Sorted Z-values of Pitx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_46120327 | 20.04 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr1_-_13730732 | 4.85 |
ENSMUST00000027071.7
|
Lactb2
|
lactamase, beta 2 |
| chr5_-_87240405 | 4.81 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr2_+_102488985 | 4.60 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_-_70405429 | 3.93 |
ENSMUST00000021179.4
|
Vmo1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr14_+_36776775 | 3.93 |
ENSMUST00000120052.2
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr3_+_118355778 | 3.49 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr3_+_97536120 | 3.33 |
ENSMUST00000107050.8
ENSMUST00000029729.15 ENSMUST00000107049.2 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr2_-_23939401 | 3.29 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr10_-_115198093 | 2.84 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr7_-_80053063 | 2.63 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_+_22958956 | 2.51 |
ENSMUST00000226571.2
ENSMUST00000114529.10 ENSMUST00000114526.9 ENSMUST00000228050.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr1_+_167426019 | 2.37 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr7_+_43361930 | 2.31 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr6_-_138013901 | 2.29 |
ENSMUST00000150278.3
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr3_+_118355811 | 2.27 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr10_-_115197775 | 2.13 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr7_+_30252687 | 2.12 |
ENSMUST00000044048.8
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr14_-_31362835 | 2.07 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr18_-_20192535 | 2.04 |
ENSMUST00000075214.9
ENSMUST00000039247.11 |
Dsc2
|
desmocollin 2 |
| chr5_+_87148697 | 1.93 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chrX_+_10118544 | 1.87 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr14_+_28740162 | 1.85 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr15_+_4404965 | 1.80 |
ENSMUST00000061925.5
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr2_+_22958179 | 1.77 |
ENSMUST00000227663.2
ENSMUST00000028121.15 ENSMUST00000227809.2 ENSMUST00000144088.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr17_-_57366795 | 1.77 |
ENSMUST00000040280.14
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chrX_+_10118600 | 1.68 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr3_+_94217396 | 1.68 |
ENSMUST00000049822.10
|
Them4
|
thioesterase superfamily member 4 |
| chr19_-_32689687 | 1.64 |
ENSMUST00000237752.2
ENSMUST00000235412.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr1_+_167425953 | 1.64 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr2_-_38816229 | 1.64 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr7_+_107166925 | 1.59 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr11_-_51647290 | 1.56 |
ENSMUST00000109097.9
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr11_-_29197222 | 1.52 |
ENSMUST00000020754.10
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr14_-_64654397 | 1.50 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr6_-_124817155 | 1.41 |
ENSMUST00000024206.6
|
Gnb3
|
guanine nucleotide binding protein (G protein), beta 3 |
| chr18_-_3299536 | 1.37 |
ENSMUST00000129435.8
ENSMUST00000122958.8 |
Crem
|
cAMP responsive element modulator |
| chr10_+_80165787 | 1.35 |
ENSMUST00000105358.8
ENSMUST00000105357.2 ENSMUST00000105354.8 ENSMUST00000105355.8 |
Reep6
|
receptor accessory protein 6 |
| chr6_+_3993774 | 1.35 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr6_-_93890237 | 1.27 |
ENSMUST00000204167.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr9_-_108444561 | 1.24 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr6_-_29380423 | 1.20 |
ENSMUST00000147483.3
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr1_-_173195236 | 1.19 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr17_-_85397627 | 1.18 |
ENSMUST00000072406.5
ENSMUST00000171795.9 ENSMUST00000234676.2 |
Prepl
|
prolyl endopeptidase-like |
| chr13_+_113171645 | 1.18 |
ENSMUST00000180543.8
ENSMUST00000181568.8 ENSMUST00000109244.9 ENSMUST00000181117.8 ENSMUST00000181741.2 |
Cdc20b
|
cell division cycle 20B |
| chr10_+_80165961 | 1.15 |
ENSMUST00000186864.7
ENSMUST00000040081.7 |
Reep6
|
receptor accessory protein 6 |
| chr4_+_84802592 | 1.15 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr9_-_54408780 | 1.14 |
ENSMUST00000118600.8
ENSMUST00000118163.8 |
Dmxl2
|
Dmx-like 2 |
| chr9_+_30941924 | 1.14 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr6_+_119213541 | 1.14 |
ENSMUST00000186622.3
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr14_+_26761023 | 1.13 |
ENSMUST00000223942.2
|
Il17rd
|
interleukin 17 receptor D |
| chr3_-_33137165 | 1.11 |
ENSMUST00000078226.10
ENSMUST00000108224.8 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr9_-_108443916 | 1.11 |
ENSMUST00000194381.2
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr15_-_79138929 | 1.03 |
ENSMUST00000039752.4
|
Slc16a8
|
solute carrier family 16 (monocarboxylic acid transporters), member 8 |
| chr5_-_151051000 | 1.03 |
ENSMUST00000202111.4
|
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr1_+_177272215 | 1.03 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr19_+_4189786 | 1.01 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr10_+_128061699 | 0.99 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr6_+_119213468 | 0.98 |
ENSMUST00000238905.2
ENSMUST00000037434.13 |
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr2_-_145776934 | 0.97 |
ENSMUST00000001818.5
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr6_-_29380467 | 0.96 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr2_+_153334710 | 0.96 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr11_+_66915969 | 0.94 |
ENSMUST00000079077.12
ENSMUST00000061786.6 |
Tmem220
|
transmembrane protein 220 |
| chr18_+_75138910 | 0.94 |
ENSMUST00000040284.6
ENSMUST00000236421.2 ENSMUST00000237263.2 |
BC031181
|
cDNA sequence BC031181 |
| chr15_-_101801351 | 0.93 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr10_+_69761630 | 0.91 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr11_-_43792013 | 0.90 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr18_-_32271224 | 0.89 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chr7_-_109092834 | 0.88 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chrM_+_8603 | 0.88 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr18_-_3299452 | 0.88 |
ENSMUST00000126578.8
|
Crem
|
cAMP responsive element modulator |
| chr6_-_122317156 | 0.87 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr9_+_58041818 | 0.87 |
ENSMUST00000128378.8
ENSMUST00000150820.8 ENSMUST00000167479.8 |
Stra6
|
stimulated by retinoic acid gene 6 |
| chr6_+_136931519 | 0.86 |
ENSMUST00000137768.2
|
Pde6h
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr9_+_58042087 | 0.84 |
ENSMUST00000134450.2
|
Stra6
|
stimulated by retinoic acid gene 6 |
| chr17_-_29768586 | 0.83 |
ENSMUST00000234305.2
ENSMUST00000234648.2 ENSMUST00000234979.2 |
Gm17657
Tmem217
|
predicted gene, 17657 transmembrane protein 217 |
| chr13_-_116446166 | 0.82 |
ENSMUST00000036060.13
|
Isl1
|
ISL1 transcription factor, LIM/homeodomain |
| chr11_+_87938128 | 0.80 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr5_-_92583078 | 0.80 |
ENSMUST00000038514.15
|
Nup54
|
nucleoporin 54 |
| chr6_-_93890659 | 0.79 |
ENSMUST00000093769.8
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr18_-_46730547 | 0.79 |
ENSMUST00000151189.2
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr10_+_85763545 | 0.79 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr11_+_116547932 | 0.78 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chr7_-_4527228 | 0.78 |
ENSMUST00000154913.2
|
Tnni3
|
troponin I, cardiac 3 |
| chr2_-_22930149 | 0.78 |
ENSMUST00000091394.13
ENSMUST00000093171.13 |
Abi1
|
abl interactor 1 |
| chr14_-_118289557 | 0.77 |
ENSMUST00000022725.4
|
Dct
|
dopachrome tautomerase |
| chr9_+_50528813 | 0.77 |
ENSMUST00000141366.8
|
Pih1d2
|
PIH1 domain containing 2 |
| chr7_-_98305986 | 0.76 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr11_-_98040377 | 0.75 |
ENSMUST00000103143.10
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr11_-_100012384 | 0.75 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr3_-_113423774 | 0.75 |
ENSMUST00000092154.10
ENSMUST00000106536.8 ENSMUST00000106535.2 |
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr7_-_127324788 | 0.75 |
ENSMUST00000076091.4
|
Ctf2
|
cardiotrophin 2 |
| chr10_+_69761597 | 0.73 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr8_-_106863521 | 0.73 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr5_+_8472831 | 0.73 |
ENSMUST00000066921.10
ENSMUST00000170496.6 |
Slc25a40
|
solute carrier family 25, member 40 |
| chr17_-_28584117 | 0.73 |
ENSMUST00000233420.2
|
Tulp1
|
tubby like protein 1 |
| chrX_+_139565319 | 0.72 |
ENSMUST00000128809.2
|
Mid2
|
midline 2 |
| chr2_-_101459303 | 0.71 |
ENSMUST00000111231.10
|
Iftap
|
intraflagellar transport associated protein |
| chr9_+_32135540 | 0.70 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_+_80447389 | 0.70 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr14_-_70666513 | 0.68 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr19_-_4189603 | 0.68 |
ENSMUST00000025761.8
|
Cabp4
|
calcium binding protein 4 |
| chr11_-_100026754 | 0.68 |
ENSMUST00000107411.3
|
Krt15
|
keratin 15 |
| chr4_-_155095441 | 0.68 |
ENSMUST00000105631.9
ENSMUST00000139976.9 ENSMUST00000145662.9 |
Plch2
|
phospholipase C, eta 2 |
| chr7_-_4517559 | 0.67 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chrX_+_7594670 | 0.67 |
ENSMUST00000033489.8
|
Praf2
|
PRA1 domain family 2 |
| chr2_-_101459274 | 0.66 |
ENSMUST00000099682.9
|
Iftap
|
intraflagellar transport associated protein |
| chr2_+_69500444 | 0.66 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr13_+_74269554 | 0.65 |
ENSMUST00000036208.7
ENSMUST00000225423.2 ENSMUST00000221703.2 |
Slc9a3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| chr4_-_139974062 | 0.64 |
ENSMUST00000039331.9
|
Igsf21
|
immunoglobulin superfamily, member 21 |
| chr7_+_75879603 | 0.64 |
ENSMUST00000156166.8
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr1_-_171050004 | 0.63 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr10_-_22025169 | 0.63 |
ENSMUST00000179054.8
ENSMUST00000069372.7 |
E030030I06Rik
|
RIKEN cDNA E030030I06 gene |
| chr6_+_122684448 | 0.63 |
ENSMUST00000112581.8
ENSMUST00000112580.8 ENSMUST00000012540.5 |
Nanog
|
Nanog homeobox |
| chr3_+_30800474 | 0.63 |
ENSMUST00000108262.10
|
Samd7
|
sterile alpha motif domain containing 7 |
| chr1_-_160405464 | 0.63 |
ENSMUST00000238289.2
|
Gpr52
|
G protein-coupled receptor 52 |
| chrX_+_7775781 | 0.62 |
ENSMUST00000086274.3
|
Gm10490
|
predicted gene 10490 |
| chr10_-_80165257 | 0.61 |
ENSMUST00000020340.15
|
Pcsk4
|
proprotein convertase subtilisin/kexin type 4 |
| chr11_+_49170977 | 0.61 |
ENSMUST00000078932.2
|
Olfr1393
|
olfactory receptor 1393 |
| chr10_-_53255959 | 0.61 |
ENSMUST00000220443.2
|
Cep85l
|
centrosomal protein 85-like |
| chr17_-_29768531 | 0.61 |
ENSMUST00000168339.3
ENSMUST00000114683.10 ENSMUST00000234620.2 |
Tmem217
|
transmembrane protein 217 |
| chr16_-_17732923 | 0.60 |
ENSMUST00000012279.6
ENSMUST00000232493.2 |
Gsc2
|
goosecoid homebox 2 |
| chr6_+_104469751 | 0.60 |
ENSMUST00000161446.2
ENSMUST00000161070.8 ENSMUST00000089215.12 |
Cntn6
|
contactin 6 |
| chr6_-_59403264 | 0.60 |
ENSMUST00000051065.6
|
Gprin3
|
GPRIN family member 3 |
| chr6_-_93890520 | 0.60 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_+_69761314 | 0.60 |
ENSMUST00000182692.8
ENSMUST00000092433.12 |
Ank3
|
ankyrin 3, epithelial |
| chrX_+_41239872 | 0.58 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr9_+_32135781 | 0.58 |
ENSMUST00000183121.2
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr2_+_136733421 | 0.58 |
ENSMUST00000141463.8
|
Slx4ip
|
SLX4 interacting protein |
| chr14_-_57049173 | 0.58 |
ENSMUST00000039812.16
ENSMUST00000111285.9 |
Zmym5
|
zinc finger, MYM-type 5 |
| chr3_-_84178089 | 0.58 |
ENSMUST00000054990.11
|
Trim2
|
tripartite motif-containing 2 |
| chr1_-_13442658 | 0.57 |
ENSMUST00000081713.11
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr11_+_60956624 | 0.56 |
ENSMUST00000041944.9
ENSMUST00000108717.3 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr7_-_4517608 | 0.56 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr8_-_11728727 | 0.56 |
ENSMUST00000033906.11
|
1700016D06Rik
|
RIKEN cDNA 1700016D06 gene |
| chr11_+_70350963 | 0.55 |
ENSMUST00000126105.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr3_-_123484499 | 0.55 |
ENSMUST00000154668.8
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr3_+_41517790 | 0.55 |
ENSMUST00000163764.8
|
Jade1
|
jade family PHD finger 1 |
| chr17_+_35188888 | 0.54 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr7_-_98305737 | 0.54 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr1_-_171050077 | 0.53 |
ENSMUST00000005817.9
|
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr4_+_136349924 | 0.53 |
ENSMUST00000178843.3
|
Lactbl1
|
lactamase, beta-like 1 |
| chr2_-_9888804 | 0.52 |
ENSMUST00000114915.3
|
9230102O04Rik
|
RIKEN cDNA 9230102O04 gene |
| chr18_+_46730765 | 0.52 |
ENSMUST00000238168.2
ENSMUST00000078079.11 ENSMUST00000168382.2 ENSMUST00000235849.2 ENSMUST00000235973.2 ENSMUST00000235455.2 ENSMUST00000237478.2 |
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr6_+_15727798 | 0.52 |
ENSMUST00000128849.3
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr17_+_12803019 | 0.52 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr6_-_136150491 | 0.52 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr14_-_57048921 | 0.51 |
ENSMUST00000173954.3
|
Zmym5
|
zinc finger, MYM-type 5 |
| chrX_-_42256694 | 0.51 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr12_+_8971603 | 0.51 |
ENSMUST00000020909.4
|
Laptm4a
|
lysosomal-associated protein transmembrane 4A |
| chr6_+_115111872 | 0.51 |
ENSMUST00000009538.12
ENSMUST00000203450.2 |
Syn2
|
synapsin II |
| chr10_-_107330580 | 0.51 |
ENSMUST00000044210.5
|
Myf6
|
myogenic factor 6 |
| chr9_-_51076724 | 0.50 |
ENSMUST00000210433.2
|
Gm32742
|
predicted gene, 32742 |
| chr11_+_23206565 | 0.50 |
ENSMUST00000136235.2
|
Xpo1
|
exportin 1 |
| chr9_+_44013019 | 0.50 |
ENSMUST00000034654.9
ENSMUST00000206308.2 ENSMUST00000161381.8 |
Mfrp
|
membrane frizzled-related protein |
| chr11_+_115225557 | 0.49 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr4_+_85972125 | 0.49 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr9_+_107217786 | 0.48 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr9_+_32305259 | 0.48 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr10_-_102864904 | 0.48 |
ENSMUST00000167156.9
|
Alx1
|
ALX homeobox 1 |
| chr6_+_97906760 | 0.47 |
ENSMUST00000101123.10
|
Mitf
|
melanogenesis associated transcription factor |
| chr4_-_42661893 | 0.47 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr19_-_12742811 | 0.47 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr4_-_139560831 | 0.47 |
ENSMUST00000030508.14
|
Pax7
|
paired box 7 |
| chr4_-_109333866 | 0.47 |
ENSMUST00000030284.10
|
Rnf11
|
ring finger protein 11 |
| chr3_+_128993609 | 0.46 |
ENSMUST00000173645.4
|
Pitx2
|
paired-like homeodomain transcription factor 2 |
| chr10_+_69761784 | 0.46 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr18_+_73996743 | 0.46 |
ENSMUST00000134847.2
|
Mro
|
maestro |
| chr7_-_15613769 | 0.46 |
ENSMUST00000172758.3
ENSMUST00000044434.13 |
Crx
|
cone-rod homeobox |
| chr16_-_10131804 | 0.46 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2 |
| chr11_-_69127848 | 0.46 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr17_-_28584150 | 0.46 |
ENSMUST00000114794.2
|
Tulp1
|
tubby like protein 1 |
| chr9_+_65538352 | 0.46 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr17_-_28584182 | 0.46 |
ENSMUST00000041819.14
|
Tulp1
|
tubby like protein 1 |
| chr1_-_169938298 | 0.46 |
ENSMUST00000192312.6
|
Ddr2
|
discoidin domain receptor family, member 2 |
| chr9_-_86762450 | 0.46 |
ENSMUST00000191290.3
|
Snap91
|
synaptosomal-associated protein 91 |
| chr11_+_67586140 | 0.46 |
ENSMUST00000021290.2
|
Rcvrn
|
recoverin |
| chr12_+_36042899 | 0.45 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr13_+_96884780 | 0.45 |
ENSMUST00000208758.2
|
Ankrd31
|
ankyrin repeat domain 31 |
| chr11_-_97242842 | 0.44 |
ENSMUST00000093942.5
|
Gpr179
|
G protein-coupled receptor 179 |
| chr8_-_106863423 | 0.44 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr18_+_75500600 | 0.44 |
ENSMUST00000026999.10
|
Smad7
|
SMAD family member 7 |
| chr11_-_105347500 | 0.44 |
ENSMUST00000049995.10
ENSMUST00000100332.4 |
Marchf10
|
membrane associated ring-CH-type finger 10 |
| chr1_+_157286124 | 0.44 |
ENSMUST00000193791.6
ENSMUST00000046743.11 ENSMUST00000119891.7 |
Cryzl2
|
crystallin zeta like 2 |
| chr7_-_101517874 | 0.44 |
ENSMUST00000150184.2
|
Folr1
|
folate receptor 1 (adult) |
| chr5_-_137785903 | 0.43 |
ENSMUST00000196022.2
|
Mepce
|
methylphosphate capping enzyme |
| chr19_-_47907628 | 0.43 |
ENSMUST00000237029.2
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr9_-_86762467 | 0.43 |
ENSMUST00000074501.12
ENSMUST00000239074.2 ENSMUST00000098495.10 ENSMUST00000036347.13 ENSMUST00000074468.13 |
Snap91
|
synaptosomal-associated protein 91 |
| chr1_-_86317066 | 0.43 |
ENSMUST00000212058.2
|
Nmur1
|
neuromedin U receptor 1 |
| chr8_-_96033213 | 0.43 |
ENSMUST00000119870.9
ENSMUST00000093268.5 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr19_+_44744484 | 0.41 |
ENSMUST00000174490.9
|
Pax2
|
paired box 2 |
| chr5_+_77413282 | 0.41 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chr14_-_36770898 | 0.41 |
ENSMUST00000225070.2
ENSMUST00000022338.7 |
Rgr
|
retinal G protein coupled receptor |
| chr4_-_140780972 | 0.40 |
ENSMUST00000040222.14
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr3_+_135053762 | 0.40 |
ENSMUST00000159658.8
ENSMUST00000078568.12 |
Slc9b1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr17_+_57077258 | 0.40 |
ENSMUST00000168666.3
|
Prr22
|
proline rich 22 |
| chr2_-_113047313 | 0.39 |
ENSMUST00000208135.2
ENSMUST00000134358.9 |
Ryr3
|
ryanodine receptor 3 |
| chr6_+_115911302 | 0.39 |
ENSMUST00000203877.2
|
Rho
|
rhodopsin |
| chr7_+_63803663 | 0.39 |
ENSMUST00000206314.2
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr2_-_113047397 | 0.39 |
ENSMUST00000080673.13
ENSMUST00000208151.2 ENSMUST00000208290.2 |
Ryr3
|
ryanodine receptor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 1.7 | 20.0 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.2 | 3.6 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.7 | 3.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.7 | 4.6 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 2.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.4 | 1.8 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.4 | 4.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 0.9 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.3 | 2.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 0.8 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.3 | 7.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 3.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 1.7 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 2.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.8 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.2 | 4.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 1.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.2 | 0.7 | GO:0000966 | pachytene(GO:0000239) RNA 5'-end processing(GO:0000966) |
| 0.2 | 0.7 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 | 1.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.5 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 0.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.4 | GO:2000595 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.1 | 0.8 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 1.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.4 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 0.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.6 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) |
| 0.1 | 0.4 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.8 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 1.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.4 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.9 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.6 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.5 | GO:0060578 | subthalamic nucleus development(GO:0021763) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 4.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.3 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 1.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.1 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.1 | 1.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.5 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.2 | GO:0021629 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.1 | 0.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.1 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 1.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.6 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.3 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 2.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 2.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0060112 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.8 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.5 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 1.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.5 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 1.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 1.0 | GO:0010659 | cardiac muscle cell apoptotic process(GO:0010659) |
| 0.0 | 2.5 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 1.0 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.9 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.1 | GO:0061643 | chemoattraction of axon(GO:0061642) chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 1.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.5 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.7 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.3 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0044317 | rod spherule(GO:0044317) |
| 0.4 | 1.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 0.9 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.2 | 1.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 1.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 2.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.7 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 22.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.5 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 4.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 4.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 7.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 4.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 1.7 | 20.0 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.9 | 3.6 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.8 | 4.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 4.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.4 | 1.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 3.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 3.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.9 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 0.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 1.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 1.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.2 | 0.7 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 1.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 0.9 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.5 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 2.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 6.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.8 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 2.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.4 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.6 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 3.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 4.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.6 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 4.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0005138 | ciliary neurotrophic factor receptor binding(GO:0005127) interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 2.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 2.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 1.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 2.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 3.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.0 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 2.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.3 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 20.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 3.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 5.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 3.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 1.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.2 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 2.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 5.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |