Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
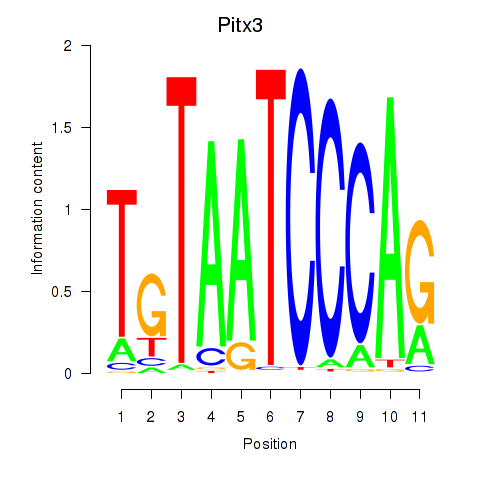
Results for Pitx3
Z-value: 0.74
Transcription factors associated with Pitx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx3
|
ENSMUSG00000025229.16 | paired-like homeodomain transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx3 | mm39_v1_chr19_-_46136765_46136825 | -0.51 | 1.4e-03 | Click! |
Activity profile of Pitx3 motif
Sorted Z-values of Pitx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_90638580 | 12.26 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr9_+_110248815 | 9.45 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr16_+_36004452 | 8.04 |
ENSMUST00000114858.2
|
Cstdc4
|
cystatin domain containing 4 |
| chr10_+_79722081 | 7.09 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr10_+_128736827 | 6.91 |
ENSMUST00000219317.2
|
Cd63
|
CD63 antigen |
| chr7_-_143013899 | 5.48 |
ENSMUST00000208137.2
ENSMUST00000207910.2 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr7_-_143014726 | 5.44 |
ENSMUST00000167912.9
ENSMUST00000037287.8 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr8_+_85628557 | 4.59 |
ENSMUST00000067060.10
ENSMUST00000239392.2 |
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chr10_+_128744689 | 4.53 |
ENSMUST00000105229.9
|
Cd63
|
CD63 antigen |
| chr3_-_36626101 | 4.31 |
ENSMUST00000029270.10
|
Ccna2
|
cyclin A2 |
| chr2_-_32278245 | 3.79 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr9_-_70328816 | 3.77 |
ENSMUST00000034742.8
|
Ccnb2
|
cyclin B2 |
| chr11_-_6470918 | 3.72 |
ENSMUST00000003459.4
|
Myo1g
|
myosin IG |
| chr15_-_78739717 | 3.66 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr6_-_88875646 | 3.58 |
ENSMUST00000058011.8
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr7_-_45173193 | 3.31 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr6_-_115739284 | 3.23 |
ENSMUST00000166254.7
ENSMUST00000170625.8 |
Tmem40
|
transmembrane protein 40 |
| chr11_+_117673107 | 3.20 |
ENSMUST00000050874.14
ENSMUST00000106334.9 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr10_-_62178453 | 3.18 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr7_-_45175570 | 3.17 |
ENSMUST00000167273.2
ENSMUST00000042105.11 |
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr17_-_35407403 | 3.05 |
ENSMUST00000097336.5
|
Lst1
|
leukocyte specific transcript 1 |
| chr11_-_116472272 | 3.04 |
ENSMUST00000082152.5
|
Ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr2_+_91376650 | 3.00 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr9_-_107863062 | 3.00 |
ENSMUST00000048568.6
|
Inka1
|
inka box actin regulator 1 |
| chr11_-_97886997 | 2.91 |
ENSMUST00000042971.16
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr14_-_70867588 | 2.90 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr15_-_82128888 | 2.79 |
ENSMUST00000089155.6
ENSMUST00000089157.11 |
Cenpm
|
centromere protein M |
| chr2_-_165242307 | 2.70 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr11_+_61847589 | 2.62 |
ENSMUST00000201671.4
ENSMUST00000202178.4 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr11_-_106889291 | 2.62 |
ENSMUST00000124541.8
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr7_+_28140352 | 2.54 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chrX_-_9335525 | 2.40 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr17_+_56259617 | 2.36 |
ENSMUST00000003274.8
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chrX_-_137985960 | 2.29 |
ENSMUST00000033626.15
ENSMUST00000060824.4 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr15_+_6451721 | 2.24 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr18_+_34869412 | 2.24 |
ENSMUST00000105038.3
|
Gm3550
|
predicted gene 3550 |
| chr14_+_31888061 | 2.22 |
ENSMUST00000164341.2
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chr5_-_100720063 | 2.22 |
ENSMUST00000031264.12
|
Plac8
|
placenta-specific 8 |
| chrX_-_137985979 | 2.20 |
ENSMUST00000152457.2
|
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr6_+_86605146 | 2.11 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr10_+_20223516 | 2.06 |
ENSMUST00000169712.3
ENSMUST00000217608.2 |
Mtfr2
|
mitochondrial fission regulator 2 |
| chr16_-_8610165 | 2.05 |
ENSMUST00000160405.8
|
Usp7
|
ubiquitin specific peptidase 7 |
| chr16_-_36154692 | 2.04 |
ENSMUST00000114850.3
|
Cstdc6
|
cystatin domain containing 6 |
| chr7_+_28140450 | 1.98 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr3_+_107137924 | 1.97 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr9_+_103182352 | 1.93 |
ENSMUST00000035164.10
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chrX_-_7956682 | 1.92 |
ENSMUST00000033505.7
|
Was
|
Wiskott-Aldrich syndrome |
| chr17_+_29333116 | 1.89 |
ENSMUST00000233717.2
ENSMUST00000141239.2 |
Rab44
|
RAB44, member RAS oncogene family |
| chr7_+_140711181 | 1.89 |
ENSMUST00000026568.10
|
Ptdss2
|
phosphatidylserine synthase 2 |
| chr10_+_62782786 | 1.89 |
ENSMUST00000131422.8
|
Dna2
|
DNA replication helicase/nuclease 2 |
| chr3_-_105839980 | 1.86 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr11_+_61847622 | 1.82 |
ENSMUST00000202389.4
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr6_+_34331054 | 1.80 |
ENSMUST00000038406.7
|
Akr1b8
|
aldo-keto reductase family 1, member B8 |
| chr18_+_35932894 | 1.79 |
ENSMUST00000236484.2
ENSMUST00000236550.2 ENSMUST00000235919.2 |
Ube2d2a
Gm50371
|
ubiquitin-conjugating enzyme E2D 2A predicted gene, 50371 |
| chr8_-_124709859 | 1.79 |
ENSMUST00000075578.7
|
Abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr17_-_25946370 | 1.79 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr17_-_26163906 | 1.79 |
ENSMUST00000085027.4
ENSMUST00000237570.2 |
Nhlrc4
Pigq
|
NHL repeat containing 4 phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr6_-_113412708 | 1.76 |
ENSMUST00000032416.11
ENSMUST00000113089.8 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr7_-_133304244 | 1.76 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chr7_-_19410749 | 1.71 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr17_+_6697511 | 1.69 |
ENSMUST00000179569.3
|
Dynlt1b
|
dynein light chain Tctex-type 1B |
| chr14_-_47655621 | 1.68 |
ENSMUST00000180299.8
|
Dlgap5
|
DLG associated protein 5 |
| chr17_-_6923299 | 1.65 |
ENSMUST00000179554.3
|
Dynlt1f
|
dynein light chain Tctex-type 1F |
| chr3_+_129672205 | 1.65 |
ENSMUST00000029629.15
|
Pla2g12a
|
phospholipase A2, group XIIA |
| chr7_-_6699422 | 1.62 |
ENSMUST00000122432.4
ENSMUST00000002336.16 |
Zim1
|
zinc finger, imprinted 1 |
| chr15_-_99268311 | 1.60 |
ENSMUST00000081224.14
ENSMUST00000120633.2 ENSMUST00000088233.13 |
Fmnl3
|
formin-like 3 |
| chr9_+_72345267 | 1.60 |
ENSMUST00000183809.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr17_+_36172210 | 1.59 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr4_-_149221998 | 1.54 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr17_+_29712008 | 1.54 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chr2_-_34645241 | 1.52 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr2_-_121304521 | 1.50 |
ENSMUST00000056732.4
|
Mfap1b
|
microfibrillar-associated protein 1B |
| chr11_+_32155415 | 1.48 |
ENSMUST00000039601.10
ENSMUST00000149043.3 |
Snrnp25
|
small nuclear ribonucleoprotein 25 (U11/U12) |
| chr9_+_118335294 | 1.48 |
ENSMUST00000084820.6
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr4_-_116485118 | 1.43 |
ENSMUST00000030456.14
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr4_+_132903646 | 1.42 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr7_-_100164007 | 1.42 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr17_+_6869070 | 1.42 |
ENSMUST00000092966.5
|
Dynlt1c
|
dynein light chain Tctex-type 1C |
| chr15_-_82128538 | 1.41 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr1_-_153363354 | 1.41 |
ENSMUST00000186380.7
ENSMUST00000188345.2 ENSMUST00000042141.12 |
Dhx9
|
DEAH (Asp-Glu-Ala-His) box polypeptide 9 |
| chr7_-_24997393 | 1.41 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr6_-_143045731 | 1.40 |
ENSMUST00000203673.3
ENSMUST00000203187.3 ENSMUST00000171349.8 ENSMUST00000087485.7 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr9_-_107512511 | 1.39 |
ENSMUST00000192615.6
ENSMUST00000192837.2 ENSMUST00000193876.2 |
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr5_-_137609634 | 1.38 |
ENSMUST00000054564.13
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr7_+_16726633 | 1.38 |
ENSMUST00000094805.5
|
Ccdc8
|
coiled-coil domain containing 8 |
| chr11_-_120238917 | 1.37 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr5_+_110478558 | 1.37 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr7_-_30119227 | 1.37 |
ENSMUST00000208740.2
ENSMUST00000075062.5 |
Hcst
|
hematopoietic cell signal transducer |
| chr16_+_31241085 | 1.35 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr1_-_153425791 | 1.35 |
ENSMUST00000041874.9
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr4_+_154096188 | 1.34 |
ENSMUST00000169622.8
ENSMUST00000030894.15 |
Lrrc47
|
leucine rich repeat containing 47 |
| chr10_+_128073900 | 1.34 |
ENSMUST00000105245.3
|
Timeless
|
timeless circadian clock 1 |
| chr15_+_97682210 | 1.33 |
ENSMUST00000117892.2
ENSMUST00000229084.2 |
Slc48a1
|
solute carrier family 48 (heme transporter), member 1 |
| chr7_-_19504446 | 1.33 |
ENSMUST00000003061.14
|
Bcam
|
basal cell adhesion molecule |
| chr4_-_149222057 | 1.32 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr9_+_15217548 | 1.30 |
ENSMUST00000215124.2
ENSMUST00000164079.9 ENSMUST00000216109.2 |
Taf1d
|
TATA-box binding protein associated factor, RNA polymerase I, D |
| chr5_-_137609691 | 1.29 |
ENSMUST00000031731.14
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr8_+_61343444 | 1.29 |
ENSMUST00000146863.2
ENSMUST00000037190.15 |
Hpf1
|
histone PARylation factor 1 |
| chr9_-_107512566 | 1.28 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr4_-_117013396 | 1.27 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
| chr5_+_117271632 | 1.27 |
ENSMUST00000179276.8
ENSMUST00000092889.12 ENSMUST00000145640.8 |
Taok3
|
TAO kinase 3 |
| chr7_-_24997291 | 1.27 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr8_-_84467798 | 1.24 |
ENSMUST00000075843.13
ENSMUST00000109802.3 ENSMUST00000166939.8 ENSMUST00000002964.14 |
Adgre5
|
adhesion G protein-coupled receptor E5 |
| chr10_-_93725619 | 1.23 |
ENSMUST00000181091.8
ENSMUST00000181217.8 ENSMUST00000047910.15 ENSMUST00000180688.2 |
Metap2
|
methionine aminopeptidase 2 |
| chr18_-_34784746 | 1.23 |
ENSMUST00000025228.12
ENSMUST00000133181.2 |
Cdc23
|
CDC23 cell division cycle 23 |
| chr10_-_62628008 | 1.22 |
ENSMUST00000217768.2
ENSMUST00000020268.7 ENSMUST00000218946.2 ENSMUST00000219527.2 |
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr11_-_117673008 | 1.22 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr1_+_92900834 | 1.21 |
ENSMUST00000186298.7
ENSMUST00000027489.9 |
Gpr35
|
G protein-coupled receptor 35 |
| chr7_+_19024994 | 1.21 |
ENSMUST00000108468.5
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr1_+_85538554 | 1.21 |
ENSMUST00000162925.2
|
Sp140
|
Sp140 nuclear body protein |
| chr4_+_156194427 | 1.21 |
ENSMUST00000072554.13
ENSMUST00000169550.8 ENSMUST00000105576.2 |
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene |
| chr12_-_28685849 | 1.20 |
ENSMUST00000221871.2
|
Rps7
|
ribosomal protein S7 |
| chr5_-_129856267 | 1.19 |
ENSMUST00000201394.4
|
Psph
|
phosphoserine phosphatase |
| chr7_-_33867940 | 1.19 |
ENSMUST00000102746.11
|
Uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr5_-_100126773 | 1.19 |
ENSMUST00000112939.10
ENSMUST00000171786.8 ENSMUST00000072750.13 ENSMUST00000019128.15 ENSMUST00000172361.8 |
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr2_+_127050281 | 1.17 |
ENSMUST00000103220.4
|
Snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr7_+_27967222 | 1.16 |
ENSMUST00000059596.8
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr19_+_8870362 | 1.15 |
ENSMUST00000096249.7
|
Ints5
|
integrator complex subunit 5 |
| chr7_-_15701395 | 1.15 |
ENSMUST00000144956.2
ENSMUST00000098799.5 |
Ehd2
|
EH-domain containing 2 |
| chr7_+_12631727 | 1.15 |
ENSMUST00000055528.11
ENSMUST00000117189.2 ENSMUST00000120809.2 ENSMUST00000119989.3 |
Zscan22
|
zinc finger and SCAN domain containing 22 |
| chr3_+_89979948 | 1.15 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr8_+_95393228 | 1.14 |
ENSMUST00000034228.16
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr5_+_108048367 | 1.13 |
ENSMUST00000082223.13
|
Rpl5
|
ribosomal protein L5 |
| chr2_+_128942919 | 1.13 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr7_+_127503251 | 1.11 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr6_+_40941688 | 1.11 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr8_-_120296734 | 1.11 |
ENSMUST00000212517.2
|
Hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr8_+_72994152 | 1.11 |
ENSMUST00000126885.2
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr11_-_103247150 | 1.11 |
ENSMUST00000136491.3
ENSMUST00000107023.3 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr11_-_121120052 | 1.10 |
ENSMUST00000169393.8
ENSMUST00000106115.8 ENSMUST00000038709.14 ENSMUST00000147490.6 |
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr2_-_163486998 | 1.10 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr17_-_33932346 | 1.10 |
ENSMUST00000173392.2
|
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr5_-_116162415 | 1.09 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr2_-_5900130 | 1.08 |
ENSMUST00000026926.5
ENSMUST00000193792.6 ENSMUST00000102981.10 |
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae) |
| chr5_+_130058119 | 1.07 |
ENSMUST00000026608.11
ENSMUST00000202163.4 ENSMUST00000202756.2 |
Crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr11_+_68906737 | 1.07 |
ENSMUST00000021278.14
ENSMUST00000161455.8 ENSMUST00000116359.3 |
Ctc1
|
CTS telomere maintenance complex component 1 |
| chr14_-_99231754 | 1.07 |
ENSMUST00000081987.5
|
Rpl36a-ps1
|
ribosomal protein L36A, pseudogene 1 |
| chr5_-_92457862 | 1.07 |
ENSMUST00000031364.5
|
Sdad1
|
SDA1 domain containing 1 |
| chr12_-_28685913 | 1.06 |
ENSMUST00000074267.5
|
Rps7
|
ribosomal protein S7 |
| chr6_+_42326714 | 1.06 |
ENSMUST00000203846.3
|
Zyx
|
zyxin |
| chr10_+_7543260 | 1.06 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr8_-_11685726 | 1.06 |
ENSMUST00000033905.13
ENSMUST00000169782.3 |
Ankrd10
|
ankyrin repeat domain 10 |
| chr6_-_131270136 | 1.06 |
ENSMUST00000032307.12
|
Magohb
|
mago homolog B, exon junction complex core component |
| chr18_-_74340842 | 1.05 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr17_+_36172235 | 1.05 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr10_-_40178182 | 1.05 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr9_-_78396407 | 1.04 |
ENSMUST00000154207.8
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_-_115918784 | 1.04 |
ENSMUST00000106454.8
|
H3f3b
|
H3.3 histone B |
| chr14_+_31881822 | 1.04 |
ENSMUST00000163336.8
ENSMUST00000169722.8 ENSMUST00000168385.8 |
Ncoa4
|
nuclear receptor coactivator 4 |
| chr9_-_114811807 | 1.02 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr11_+_117673198 | 1.02 |
ENSMUST00000117781.8
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr16_+_17051423 | 1.01 |
ENSMUST00000090190.14
ENSMUST00000232082.2 ENSMUST00000232426.2 |
Hic2
Gm49573
|
hypermethylated in cancer 2 predicted gene, 49573 |
| chr19_-_17316906 | 1.00 |
ENSMUST00000169897.2
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr14_+_47710005 | 1.00 |
ENSMUST00000043112.9
ENSMUST00000163324.8 ENSMUST00000228668.2 ENSMUST00000168833.9 |
Fbxo34
|
F-box protein 34 |
| chr10_+_58230203 | 0.99 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr9_+_21231994 | 0.98 |
ENSMUST00000217461.2
|
Slc44a2
|
solute carrier family 44, member 2 |
| chr12_+_83572774 | 0.98 |
ENSMUST00000223291.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr12_-_69205882 | 0.98 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr2_+_128942900 | 0.97 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr7_-_4448631 | 0.97 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr7_-_117715394 | 0.97 |
ENSMUST00000131374.8
|
Rps15a
|
ribosomal protein S15A |
| chr7_-_47178610 | 0.97 |
ENSMUST00000172559.2
|
Mrgpra2b
|
MAS-related GPR, member A2B |
| chr10_+_79832313 | 0.96 |
ENSMUST00000132517.8
|
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr6_+_42326934 | 0.96 |
ENSMUST00000203401.3
ENSMUST00000164375.4 |
Zyx
|
zyxin |
| chr15_+_102391614 | 0.96 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr1_-_55066629 | 0.95 |
ENSMUST00000027127.14
|
Sf3b1
|
splicing factor 3b, subunit 1 |
| chr19_-_32188413 | 0.95 |
ENSMUST00000151289.9
|
Sgms1
|
sphingomyelin synthase 1 |
| chr16_+_16844217 | 0.95 |
ENSMUST00000232067.2
|
Mapk1
|
mitogen-activated protein kinase 1 |
| chr5_-_129856237 | 0.94 |
ENSMUST00000118268.9
|
Psph
|
phosphoserine phosphatase |
| chr16_-_4536992 | 0.94 |
ENSMUST00000115851.10
|
Nmral1
|
NmrA-like family domain containing 1 |
| chr13_-_38842967 | 0.94 |
ENSMUST00000001757.9
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr16_-_4536943 | 0.94 |
ENSMUST00000120056.8
ENSMUST00000074970.8 |
Nmral1
|
NmrA-like family domain containing 1 |
| chr11_+_96822213 | 0.93 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr5_-_36853281 | 0.92 |
ENSMUST00000031091.13
ENSMUST00000140653.2 |
D5Ertd579e
|
DNA segment, Chr 5, ERATO Doi 579, expressed |
| chr5_-_92457753 | 0.92 |
ENSMUST00000201143.2
|
Sdad1
|
SDA1 domain containing 1 |
| chr17_-_66175046 | 0.91 |
ENSMUST00000233399.2
|
Ralbp1
|
ralA binding protein 1 |
| chr3_+_30910089 | 0.91 |
ENSMUST00000108261.8
ENSMUST00000108259.8 ENSMUST00000166278.7 ENSMUST00000046748.13 ENSMUST00000194979.6 |
Gpr160
|
G protein-coupled receptor 160 |
| chr12_-_79027531 | 0.91 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr16_-_22084700 | 0.90 |
ENSMUST00000161286.8
|
Tra2b
|
transformer 2 beta |
| chr10_+_58230183 | 0.90 |
ENSMUST00000020077.11
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_+_102387053 | 0.89 |
ENSMUST00000230682.2
ENSMUST00000231085.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr5_-_134485430 | 0.89 |
ENSMUST00000200944.4
ENSMUST00000202280.4 ENSMUST00000074114.12 ENSMUST00000100654.10 ENSMUST00000100650.10 ENSMUST00000111245.9 ENSMUST00000202554.4 ENSMUST00000073161.12 ENSMUST00000100652.10 ENSMUST00000171794.9 ENSMUST00000167084.9 |
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr9_+_69902697 | 0.88 |
ENSMUST00000165389.8
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr9_+_64868127 | 0.87 |
ENSMUST00000170517.9
ENSMUST00000037504.7 |
Ints14
|
integrator complex subunit 14 |
| chr11_-_78074377 | 0.87 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr11_-_106890195 | 0.86 |
ENSMUST00000106768.2
ENSMUST00000144834.8 |
Kpna2
|
karyopherin (importin) alpha 2 |
| chr5_+_115697526 | 0.86 |
ENSMUST00000086519.12
ENSMUST00000156359.2 ENSMUST00000152976.2 |
Rplp0
|
ribosomal protein, large, P0 |
| chrX_+_162923474 | 0.86 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr3_+_95434093 | 0.86 |
ENSMUST00000015667.9
ENSMUST00000116304.3 |
Ctss
|
cathepsin S |
| chr13_-_101829132 | 0.86 |
ENSMUST00000035532.13
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr7_-_126831803 | 0.85 |
ENSMUST00000133913.8
|
Septin1
|
septin 1 |
| chr8_+_26210484 | 0.85 |
ENSMUST00000210629.2
|
Plpp5
|
phospholipid phosphatase 5 |
| chr2_+_31985528 | 0.84 |
ENSMUST00000057423.6
ENSMUST00000140762.2 |
Plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr11_+_88861073 | 0.82 |
ENSMUST00000107898.8
|
Coil
|
coilin |
| chr15_+_99600149 | 0.81 |
ENSMUST00000229236.2
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr6_+_42326760 | 0.81 |
ENSMUST00000203652.3
ENSMUST00000070635.13 |
Zyx
|
zyxin |
| chr6_-_120334382 | 0.81 |
ENSMUST00000032283.12
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr5_+_66018555 | 0.80 |
ENSMUST00000031106.8
|
Rhoh
|
ras homolog family member H |
| chr9_+_15217503 | 0.80 |
ENSMUST00000034415.6
|
Taf1d
|
TATA-box binding protein associated factor, RNA polymerase I, D |
| chr15_+_95698574 | 0.79 |
ENSMUST00000226793.2
|
Ano6
|
anoctamin 6 |
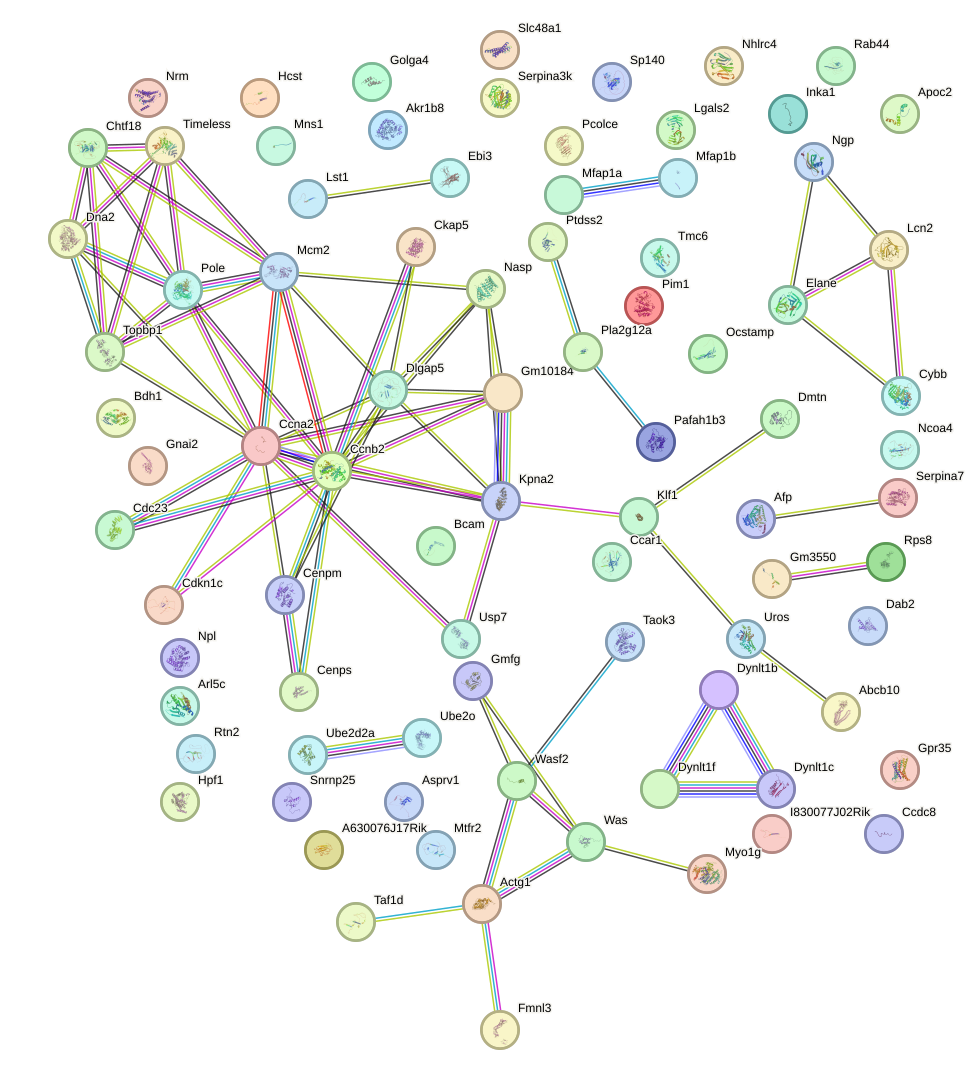
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) neutrophil mediated killing of fungus(GO:0070947) |
| 2.4 | 9.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 2.2 | 6.5 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 1.9 | 11.4 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 1.5 | 4.5 | GO:0071846 | actin filament debranching(GO:0071846) |
| 1.5 | 4.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 1.3 | 3.8 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.0 | 10.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.7 | 12.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.7 | 2.9 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.7 | 2.7 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.6 | 1.9 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.6 | 1.8 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.6 | 1.8 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.5 | 2.1 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.5 | 3.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 2.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.5 | 1.9 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.5 | 1.4 | GO:0006272 | leading strand elongation(GO:0006272) DNA replication proofreading(GO:0045004) |
| 0.4 | 1.3 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.4 | 1.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.4 | 1.7 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.4 | 2.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 1.2 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.4 | 1.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.4 | 1.1 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.4 | 2.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.4 | 1.1 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.4 | 1.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 2.8 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 1.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.7 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.3 | 2.7 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.3 | 1.0 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.3 | 3.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 1.5 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.3 | 1.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 0.9 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 1.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 1.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.3 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.3 | 2.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 0.8 | GO:0002543 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.3 | 0.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 4.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.5 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.2 | 2.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 1.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 0.9 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.2 | 1.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 0.6 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 1.0 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 0.8 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 0.6 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 0.6 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.2 | 0.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 0.8 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 1.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 0.8 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 0.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 0.7 | GO:2001188 | negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.2 | 0.6 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.2 | 0.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.7 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 2.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.2 | 0.5 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.2 | 0.7 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 | 0.7 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 3.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 4.8 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.2 | 1.5 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.2 | 0.7 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.2 | 2.3 | GO:1904667 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 2.9 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 1.4 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.2 | 0.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 0.5 | GO:0046038 | GMP catabolic process(GO:0046038) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 | 0.5 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 4.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 1.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.6 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.4 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.1 | 0.4 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.1 | 1.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.9 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 1.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.6 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 0.6 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 1.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.6 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.6 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.1 | 0.1 | GO:0071029 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) |
| 0.1 | 1.1 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.4 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 2.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.6 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.4 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 5.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.4 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 2.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.1 | 0.6 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.6 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 1.1 | GO:0048757 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.8 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 1.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.2 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.1 | 0.2 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 1.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.4 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.1 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.8 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.1 | 1.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.4 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.2 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.1 | 1.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 1.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.4 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.1 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.1 | 0.4 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 1.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 0.9 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.3 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 0.5 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.5 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.4 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.4 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 1.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 3.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.0 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 2.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 1.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 3.8 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.3 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 1.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.8 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.4 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.2 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 1.3 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 2.0 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.7 | GO:1904814 | regulation of protein localization to chromosome, telomeric region(GO:1904814) positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 1.7 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 2.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.6 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0061344 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) regulation of cell adhesion involved in heart morphogenesis(GO:0061344) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 1.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.8 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 4.2 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:1901874 | negative regulation of cellular respiration(GO:1901856) regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 1.0 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.2 | GO:1904636 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) cellular response to ether(GO:0071362) interleukin-21 secretion(GO:0072619) response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 1.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.4 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 1.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.7 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 1.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 2.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.4 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.8 | 2.3 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.7 | 2.9 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.6 | 1.9 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.5 | 1.5 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 2.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.4 | 6.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 2.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.4 | 4.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.3 | 1.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.3 | 3.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 1.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 1.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 0.8 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.2 | 0.7 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.2 | 1.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 1.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 3.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 0.6 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.2 | 1.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 1.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 2.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.8 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.2 | 2.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 1.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 5.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.8 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.6 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 0.8 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 3.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.8 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.8 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 1.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 2.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 3.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.6 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 6.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 1.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 2.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.4 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 2.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.9 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.3 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 2.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.3 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 4.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 7.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.1 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 4.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 4.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 2.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 1.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 3.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.2 | GO:0030684 | preribosome(GO:0030684) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 4.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 5.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.9 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 1.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.6 | 1.9 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.5 | 10.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.5 | 3.4 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.5 | 1.4 | GO:0045142 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.4 | 1.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.4 | 3.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 1.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.4 | 2.7 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 2.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.4 | 2.8 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 1.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 4.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.0 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 1.8 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 0.7 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.2 | 3.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 0.6 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.2 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 0.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 0.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.8 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 0.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 2.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 1.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.7 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.2 | 1.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.2 | 1.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 0.9 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 6.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 1.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 1.9 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.7 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.4 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 7.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.5 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 1.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.6 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 3.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.6 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 4.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.6 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 1.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.4 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 2.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 12.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.5 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 3.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.7 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 1.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 4.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 4.3 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 3.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.5 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.8 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.6 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 2.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.2 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 2.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.4 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 1.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.7 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 7.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 4.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 1.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 3.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 3.9 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.7 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 1.2 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 4.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 2.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.4 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0070411 | transforming growth factor beta receptor activity, type I(GO:0005025) I-SMAD binding(GO:0070411) |
| 0.0 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0033797 | selenate reductase activity(GO:0033797) mercury ion binding(GO:0045340) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.4 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 4.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 13.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 3.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 4.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 7.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 7.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 3.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 0.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 7.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 3.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 1.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 6.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 2.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 13.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 11.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 9.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 4.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 2.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 3.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 9.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 2.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.2 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 3.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |