Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
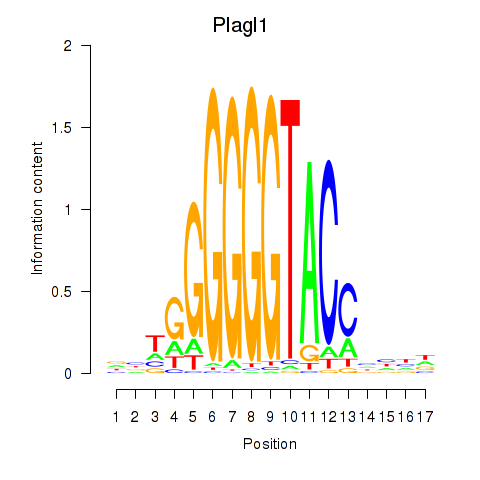
Results for Plagl1
Z-value: 0.53
Transcription factors associated with Plagl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Plagl1
|
ENSMUSG00000019817.19 | pleiomorphic adenoma gene-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plagl1 | mm39_v1_chr10_+_12936248_12936282 | -0.38 | 2.3e-02 | Click! |
Activity profile of Plagl1 motif
Sorted Z-values of Plagl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_84990360 | 2.03 |
ENSMUST00000066175.10
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr19_+_4761181 | 1.35 |
ENSMUST00000008991.8
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr7_-_99345016 | 1.33 |
ENSMUST00000107086.9
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr17_+_84990541 | 1.25 |
ENSMUST00000045714.15
ENSMUST00000171915.2 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr9_+_37466989 | 0.99 |
ENSMUST00000213126.2
|
Siae
|
sialic acid acetylesterase |
| chrX_-_74918122 | 0.78 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr5_-_87686048 | 0.73 |
ENSMUST00000031199.11
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr2_+_25318642 | 0.65 |
ENSMUST00000102919.4
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr2_+_32496990 | 0.60 |
ENSMUST00000095045.9
ENSMUST00000095044.10 ENSMUST00000126636.8 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr15_-_50753792 | 0.58 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr3_-_107925159 | 0.52 |
ENSMUST00000004140.11
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr6_-_128415640 | 0.50 |
ENSMUST00000032508.11
|
Fkbp4
|
FK506 binding protein 4 |
| chr5_+_125552878 | 0.47 |
ENSMUST00000031445.5
|
Aacs
|
acetoacetyl-CoA synthetase |
| chr5_+_137979763 | 0.47 |
ENSMUST00000035390.7
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr7_-_30062197 | 0.44 |
ENSMUST00000046351.7
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr5_+_24598633 | 0.43 |
ENSMUST00000138168.3
ENSMUST00000115077.8 |
Abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr4_+_47288057 | 0.43 |
ENSMUST00000140413.8
ENSMUST00000107731.9 |
Col15a1
|
collagen, type XV, alpha 1 |
| chr2_+_153334710 | 0.42 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr5_-_34345014 | 0.42 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr1_-_120001752 | 0.41 |
ENSMUST00000056089.8
|
Tmem37
|
transmembrane protein 37 |
| chr17_-_24863907 | 0.39 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr12_-_103409912 | 0.39 |
ENSMUST00000055071.9
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A |
| chr17_-_24863956 | 0.39 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr7_+_5018453 | 0.38 |
ENSMUST00000086349.5
ENSMUST00000207050.2 |
Zfp524
Gm44973
|
zinc finger protein 524 predicted gene 44973 |
| chr9_+_108539296 | 0.38 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr1_-_131207279 | 0.38 |
ENSMUST00000062108.10
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr6_-_128414616 | 0.37 |
ENSMUST00000151796.3
|
Fkbp4
|
FK506 binding protein 4 |
| chr9_-_58444754 | 0.37 |
ENSMUST00000213722.2
|
Cd276
|
CD276 antigen |
| chr11_+_87590720 | 0.37 |
ENSMUST00000040089.5
|
Rnf43
|
ring finger protein 43 |
| chr5_-_143255713 | 0.37 |
ENSMUST00000161448.8
|
Zfp316
|
zinc finger protein 316 |
| chr18_+_48179711 | 0.36 |
ENSMUST00000235307.2
|
Eno1b
|
enolase 1B, retrotransposed |
| chr6_-_48422447 | 0.36 |
ENSMUST00000114564.8
|
Zfp467
|
zinc finger protein 467 |
| chr5_-_135518098 | 0.35 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr7_+_142086749 | 0.34 |
ENSMUST00000038675.7
|
Mrpl23
|
mitochondrial ribosomal protein L23 |
| chr9_+_65536892 | 0.34 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr17_-_26087696 | 0.33 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr17_-_29768531 | 0.33 |
ENSMUST00000168339.3
ENSMUST00000114683.10 ENSMUST00000234620.2 |
Tmem217
|
transmembrane protein 217 |
| chr4_-_45489794 | 0.32 |
ENSMUST00000146236.8
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr11_+_67857268 | 0.31 |
ENSMUST00000021286.11
ENSMUST00000108675.2 |
Stx8
|
syntaxin 8 |
| chr15_+_76227695 | 0.31 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr1_+_74627445 | 0.30 |
ENSMUST00000113733.10
ENSMUST00000027358.11 |
Bcs1l
|
BCS1-like (yeast) |
| chr6_-_48422612 | 0.29 |
ENSMUST00000114556.2
|
Zfp467
|
zinc finger protein 467 |
| chr5_-_120605361 | 0.29 |
ENSMUST00000132916.2
|
Sdsl
|
serine dehydratase-like |
| chr8_-_14024676 | 0.27 |
ENSMUST00000210414.2
|
Tdrp
|
testis development related protein |
| chr5_-_143279378 | 0.27 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr17_-_29768586 | 0.26 |
ENSMUST00000234305.2
ENSMUST00000234648.2 ENSMUST00000234979.2 |
Gm17657
Tmem217
|
predicted gene, 17657 transmembrane protein 217 |
| chr1_+_74627506 | 0.26 |
ENSMUST00000113732.2
|
Bcs1l
|
BCS1-like (yeast) |
| chr6_-_48422759 | 0.26 |
ENSMUST00000114561.9
|
Zfp467
|
zinc finger protein 467 |
| chr4_+_47288287 | 0.25 |
ENSMUST00000146967.2
|
Col15a1
|
collagen, type XV, alpha 1 |
| chr17_+_79934096 | 0.25 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr10_-_81186222 | 0.25 |
ENSMUST00000020454.11
ENSMUST00000105324.9 ENSMUST00000154609.3 ENSMUST00000105323.8 |
Hmg20b
|
high mobility group 20B |
| chr6_-_134864731 | 0.25 |
ENSMUST00000203762.4
ENSMUST00000215088.2 ENSMUST00000066107.10 |
Gpr19
|
G protein-coupled receptor 19 |
| chr4_-_22488296 | 0.25 |
ENSMUST00000178174.3
|
Pou3f2
|
POU domain, class 3, transcription factor 2 |
| chr10_-_81186137 | 0.24 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr7_+_29003382 | 0.24 |
ENSMUST00000049977.13
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr2_-_164285097 | 0.24 |
ENSMUST00000017153.4
|
Sdc4
|
syndecan 4 |
| chrX_-_20157966 | 0.23 |
ENSMUST00000115393.3
ENSMUST00000072451.11 |
Slc9a7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7 |
| chr6_-_124689094 | 0.23 |
ENSMUST00000004379.8
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr2_-_155434487 | 0.23 |
ENSMUST00000155347.2
ENSMUST00000130881.8 ENSMUST00000079691.13 |
Gss
|
glutathione synthetase |
| chr4_+_152423075 | 0.23 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr6_-_124689001 | 0.21 |
ENSMUST00000203238.2
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chrX_+_139243012 | 0.21 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr5_+_35198853 | 0.20 |
ENSMUST00000030985.10
ENSMUST00000202573.2 |
Hgfac
|
hepatocyte growth factor activator |
| chr4_+_155334417 | 0.20 |
ENSMUST00000178473.8
ENSMUST00000105627.8 ENSMUST00000097747.9 |
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr13_-_35211060 | 0.20 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr1_+_153750081 | 0.20 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr15_-_37792635 | 0.19 |
ENSMUST00000090150.11
ENSMUST00000150453.2 |
Ncald
|
neurocalcin delta |
| chr16_-_56537545 | 0.19 |
ENSMUST00000141404.3
|
Tfg
|
Trk-fused gene |
| chr6_-_52211882 | 0.19 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chrX_+_119199956 | 0.19 |
ENSMUST00000113364.10
ENSMUST00000050239.16 ENSMUST00000113358.10 |
Pcdh11x
|
protocadherin 11 X-linked |
| chr6_+_124689241 | 0.19 |
ENSMUST00000004375.16
|
Phb2
|
prohibitin 2 |
| chr13_-_74498320 | 0.18 |
ENSMUST00000221594.2
ENSMUST00000022062.8 |
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr15_+_75881712 | 0.18 |
ENSMUST00000187868.3
|
Iqank1
|
IQ motif and ankyrin repeat containing 1 |
| chr16_-_19079594 | 0.18 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr7_+_45522196 | 0.18 |
ENSMUST00000002855.14
ENSMUST00000211716.2 |
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chrX_+_159285149 | 0.18 |
ENSMUST00000112377.8
ENSMUST00000139587.8 |
Phka2
|
phosphorylase kinase alpha 2 |
| chr18_-_61533434 | 0.18 |
ENSMUST00000063307.6
ENSMUST00000075299.13 |
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
| chr10_-_60667161 | 0.17 |
ENSMUST00000218637.2
|
Unc5b
|
unc-5 netrin receptor B |
| chr10_+_128212953 | 0.17 |
ENSMUST00000014642.10
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr16_+_10306075 | 0.16 |
ENSMUST00000023147.8
|
Ciita
|
class II transactivator |
| chr18_-_38734389 | 0.16 |
ENSMUST00000025295.8
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr16_-_56537650 | 0.16 |
ENSMUST00000128551.8
|
Tfg
|
Trk-fused gene |
| chr7_-_102148780 | 0.16 |
ENSMUST00000216116.4
|
Olfr545
|
olfactory receptor 545 |
| chr11_+_4207557 | 0.16 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr9_+_108782664 | 0.15 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr19_-_5538370 | 0.15 |
ENSMUST00000124334.8
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr1_-_135513443 | 0.14 |
ENSMUST00000067414.13
|
Nav1
|
neuron navigator 1 |
| chr8_+_94537460 | 0.14 |
ENSMUST00000034198.15
ENSMUST00000125716.8 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr3_-_96634752 | 0.14 |
ENSMUST00000154679.8
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr11_+_98632953 | 0.14 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chrX_+_159285414 | 0.14 |
ENSMUST00000033652.9
ENSMUST00000112380.9 ENSMUST00000112376.2 |
Phka2
|
phosphorylase kinase alpha 2 |
| chr2_-_152174565 | 0.14 |
ENSMUST00000028964.14
|
Rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr3_+_90011435 | 0.13 |
ENSMUST00000029548.9
ENSMUST00000200410.2 |
Nup210l
|
nucleoporin 210-like |
| chrX_-_93166964 | 0.13 |
ENSMUST00000137853.8
|
Zfx
|
zinc finger protein X-linked |
| chr2_-_65397850 | 0.13 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr6_-_91784299 | 0.13 |
ENSMUST00000159684.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr4_-_133930315 | 0.13 |
ENSMUST00000097849.3
|
Zpld2
|
zona pellucida like domain containing 2 |
| chr10_+_43097460 | 0.13 |
ENSMUST00000095725.11
|
Pdss2
|
prenyl (solanesyl) diphosphate synthase, subunit 2 |
| chr6_-_92458324 | 0.13 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr9_+_72714156 | 0.13 |
ENSMUST00000055535.9
|
Prtg
|
protogenin |
| chr9_-_117080869 | 0.13 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr6_+_42263644 | 0.12 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr17_+_46608333 | 0.12 |
ENSMUST00000188223.7
ENSMUST00000061722.13 ENSMUST00000166280.8 |
Dlk2
|
delta like non-canonical Notch ligand 2 |
| chr9_+_108782646 | 0.12 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr6_+_42263609 | 0.12 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr9_-_52591030 | 0.12 |
ENSMUST00000213937.2
|
AI593442
|
expressed sequence AI593442 |
| chr9_+_121232480 | 0.12 |
ENSMUST00000210351.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr7_+_45434755 | 0.12 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chrX_-_139714182 | 0.11 |
ENSMUST00000044179.8
|
Tex13b
|
testis expressed 13B |
| chr10_-_60667349 | 0.11 |
ENSMUST00000077925.7
|
Unc5b
|
unc-5 netrin receptor B |
| chr18_+_35987791 | 0.11 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr10_-_41592630 | 0.10 |
ENSMUST00000189488.3
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr11_+_61376257 | 0.10 |
ENSMUST00000064783.10
ENSMUST00000040522.7 |
Mfap4
|
microfibrillar-associated protein 4 |
| chr13_-_95359543 | 0.10 |
ENSMUST00000162292.8
|
Pde8b
|
phosphodiesterase 8B |
| chr6_-_91784405 | 0.10 |
ENSMUST00000162300.8
|
Grip2
|
glutamate receptor interacting protein 2 |
| chr7_+_103754080 | 0.10 |
ENSMUST00000214099.2
|
Olfr646
|
olfactory receptor 646 |
| chr15_-_43733389 | 0.09 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr2_-_65397809 | 0.09 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr11_-_78277384 | 0.09 |
ENSMUST00000108294.2
|
Foxn1
|
forkhead box N1 |
| chr12_-_103641723 | 0.09 |
ENSMUST00000095451.2
|
Serpina16
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 16 |
| chr8_+_19779592 | 0.09 |
ENSMUST00000098909.5
|
4930467E23Rik
|
RIKEN cDNA 4930467E23 gene |
| chr8_+_21154935 | 0.09 |
ENSMUST00000178438.3
|
Gm21119
|
predicted gene, 21119 |
| chr8_+_20078070 | 0.09 |
ENSMUST00000084046.6
|
Gm15319
|
predicted gene 15319 |
| chr17_+_35345292 | 0.09 |
ENSMUST00000061859.7
|
D17H6S53E
|
DNA segment, Chr 17, human D6S53E |
| chr2_-_84500951 | 0.09 |
ENSMUST00000189988.3
ENSMUST00000189636.8 ENSMUST00000102646.4 ENSMUST00000102647.11 ENSMUST00000117299.10 |
Selenoh
|
selenoprotein H |
| chrX_+_73348832 | 0.08 |
ENSMUST00000153141.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr12_-_36303394 | 0.08 |
ENSMUST00000221155.2
|
Lrrc72
|
leucine rich repeat containing 72 |
| chr18_-_89787603 | 0.08 |
ENSMUST00000097495.5
|
Dok6
|
docking protein 6 |
| chr2_-_25104628 | 0.08 |
ENSMUST00000043774.11
ENSMUST00000114363.2 |
Stpg3
|
sperm tail PG rich repeat containing 3 |
| chr4_+_59003121 | 0.08 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr12_+_16703709 | 0.07 |
ENSMUST00000221049.2
|
Ntsr2
|
neurotensin receptor 2 |
| chr10_+_19588318 | 0.07 |
ENSMUST00000020185.5
|
Il20ra
|
interleukin 20 receptor, alpha |
| chr11_+_32405367 | 0.07 |
ENSMUST00000051053.5
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr15_+_102379503 | 0.07 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr11_+_69909659 | 0.07 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr4_+_129030710 | 0.07 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr8_-_88686188 | 0.06 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr13_+_3887757 | 0.06 |
ENSMUST00000042219.6
|
Calm4
|
calmodulin 4 |
| chr15_-_102275403 | 0.06 |
ENSMUST00000229464.2
|
Sp7
|
Sp7 transcription factor 7 |
| chr18_+_35987733 | 0.06 |
ENSMUST00000235337.2
|
Cxxc5
|
CXXC finger 5 |
| chr5_-_24782465 | 0.06 |
ENSMUST00000030795.10
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr1_-_5089564 | 0.06 |
ENSMUST00000002533.15
|
Rgs20
|
regulator of G-protein signaling 20 |
| chrX_+_35592006 | 0.06 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr11_+_60360165 | 0.05 |
ENSMUST00000071880.9
ENSMUST00000081823.12 ENSMUST00000094135.9 |
Myo15
|
myosin XV |
| chr11_-_33793461 | 0.05 |
ENSMUST00000101368.9
ENSMUST00000065970.6 ENSMUST00000109340.9 |
Kcnip1
|
Kv channel-interacting protein 1 |
| chr4_+_33132502 | 0.05 |
ENSMUST00000029947.6
|
Gabrr1
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 1 |
| chr12_-_17374704 | 0.05 |
ENSMUST00000020884.16
ENSMUST00000095820.12 ENSMUST00000221129.2 ENSMUST00000127185.8 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr2_+_165345707 | 0.05 |
ENSMUST00000029196.5
|
Slc2a10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr9_+_113886208 | 0.05 |
ENSMUST00000135338.3
|
Susd5
|
sushi domain containing 5 |
| chr15_-_102274781 | 0.04 |
ENSMUST00000078508.7
|
Sp7
|
Sp7 transcription factor 7 |
| chr1_-_131025562 | 0.04 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr14_+_35816874 | 0.04 |
ENSMUST00000226305.2
|
4930474N05Rik
|
RIKEN cDNA 4930474N05 gene |
| chr19_+_5524701 | 0.04 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr6_+_41519654 | 0.04 |
ENSMUST00000103293.2
|
Trbj2-1
|
T cell receptor beta joining 2-1 |
| chr9_+_75532992 | 0.04 |
ENSMUST00000034702.6
|
Lysmd2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr4_+_102835888 | 0.04 |
ENSMUST00000223169.2
|
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr5_-_139115914 | 0.04 |
ENSMUST00000129851.8
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr15_+_102379621 | 0.03 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr11_-_119910986 | 0.03 |
ENSMUST00000134319.2
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr9_+_46184362 | 0.03 |
ENSMUST00000156440.8
ENSMUST00000114552.4 |
Zpr1
|
ZPR1 zinc finger |
| chr2_-_36899347 | 0.03 |
ENSMUST00000216437.2
|
Olfr358
|
olfactory receptor 358 |
| chr11_-_102298748 | 0.03 |
ENSMUST00000124755.2
|
Slc25a39
|
solute carrier family 25, member 39 |
| chr11_+_98632696 | 0.03 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr1_+_135980639 | 0.03 |
ENSMUST00000112064.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr9_+_39932760 | 0.01 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr6_+_67873135 | 0.01 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr6_+_41520287 | 0.01 |
ENSMUST00000103296.2
|
Trbj2-4
|
T cell receptor beta joining 2-4 |
| chr2_-_3420056 | 0.01 |
ENSMUST00000115084.8
ENSMUST00000115083.8 |
Meig1
|
meiosis expressed gene 1 |
| chr3_-_46402355 | 0.01 |
ENSMUST00000195537.2
ENSMUST00000166505.7 ENSMUST00000195436.2 |
Pabpc4l
|
poly(A) binding protein, cytoplasmic 4-like |
| chr1_-_128030148 | 0.01 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr11_+_59433554 | 0.01 |
ENSMUST00000149126.2
|
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr11_-_87788066 | 0.01 |
ENSMUST00000217095.2
ENSMUST00000215150.2 |
Olfr463
|
olfactory receptor 463 |
| chr2_-_162929732 | 0.01 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr16_-_34083200 | 0.00 |
ENSMUST00000114947.2
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr5_-_92231517 | 0.00 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr10_-_128383508 | 0.00 |
ENSMUST00000152539.8
ENSMUST00000133458.2 ENSMUST00000040572.10 |
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr6_-_112466780 | 0.00 |
ENSMUST00000053306.8
|
Oxtr
|
oxytocin receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Plagl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.3 | 1.3 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.2 | 0.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.4 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.1 | 0.6 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.4 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.4 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.4 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.7 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.2 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.2 | 1.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.2 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 0.9 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 3.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.3 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.1 | 0.4 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 1.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.5 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.5 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |