Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pou2f1
Z-value: 0.47
Transcription factors associated with Pou2f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f1
|
ENSMUSG00000026565.19 | POU domain, class 2, transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f1 | mm39_v1_chr1_-_165762469_165762605 | 0.30 | 8.0e-02 | Click! |
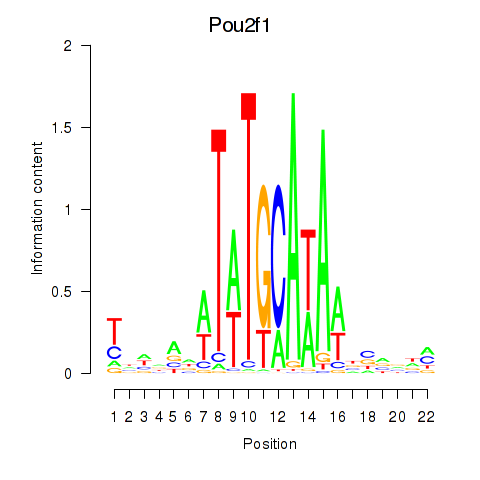
Activity profile of Pou2f1 motif
Sorted Z-values of Pou2f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41012435 | 4.73 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr6_-_41291634 | 2.98 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr7_+_13467422 | 2.90 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr7_-_13571334 | 1.92 |
ENSMUST00000108522.5
|
Sult2a1
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr7_-_13856967 | 1.72 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr4_+_134238310 | 1.09 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr4_-_132260799 | 1.04 |
ENSMUST00000152993.8
ENSMUST00000067496.7 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr12_-_76756772 | 0.90 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr4_-_46413484 | 0.88 |
ENSMUST00000071096.3
|
Hemgn
|
hemogen |
| chr10_+_75409282 | 0.87 |
ENSMUST00000006508.10
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_-_72178340 | 0.75 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr14_-_44057096 | 0.67 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr10_-_68377672 | 0.57 |
ENSMUST00000020103.9
|
Cabcoco1
|
ciliary associated calcium binding coiled-coil 1 |
| chr2_+_84564394 | 0.56 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr2_-_120867232 | 0.56 |
ENSMUST00000023987.6
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr4_-_131695135 | 0.54 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr10_+_129219952 | 0.54 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr3_+_54268523 | 0.53 |
ENSMUST00000117373.8
ENSMUST00000107985.10 ENSMUST00000073012.13 ENSMUST00000081564.13 |
Postn
|
periostin, osteoblast specific factor |
| chr1_-_144651157 | 0.50 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr1_-_131066004 | 0.49 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr14_-_44112974 | 0.47 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr10_-_62178453 | 0.46 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr4_-_149221998 | 0.44 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr1_-_128520002 | 0.42 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr16_+_36073416 | 0.42 |
ENSMUST00000063539.7
|
Csta2
|
cystatin A family member 2 |
| chr2_-_34645241 | 0.42 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr14_+_44340111 | 0.41 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr6_+_57679621 | 0.41 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr11_+_58839716 | 0.41 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chrX_+_74425990 | 0.39 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr15_-_57982705 | 0.39 |
ENSMUST00000228783.2
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr2_-_34644376 | 0.38 |
ENSMUST00000142436.2
ENSMUST00000028224.15 ENSMUST00000113099.10 |
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr15_-_81845050 | 0.37 |
ENSMUST00000071462.7
ENSMUST00000023112.12 |
Pmm1
|
phosphomannomutase 1 |
| chr13_-_22017677 | 0.37 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr13_+_23930717 | 0.36 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr4_-_132080916 | 0.35 |
ENSMUST00000155129.2
ENSMUST00000151374.2 |
Rcc1
Snhg3
|
regulator of chromosome condensation 1 small nucleolar RNA host gene 3 |
| chr7_+_24476597 | 0.35 |
ENSMUST00000038069.9
ENSMUST00000206847.2 |
Ceacam10
|
carcinoembryonic antigen-related cell adhesion molecule 10 |
| chr5_-_18093739 | 0.34 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr6_+_57679455 | 0.33 |
ENSMUST00000072954.8
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr6_+_123099615 | 0.33 |
ENSMUST00000161636.8
ENSMUST00000161365.8 |
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr15_+_99870661 | 0.32 |
ENSMUST00000100206.4
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr10_-_93727003 | 0.31 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr15_-_98851423 | 0.31 |
ENSMUST00000134214.3
|
Gm49450
|
predicted gene, 49450 |
| chr1_+_135945705 | 0.31 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr8_+_104977493 | 0.31 |
ENSMUST00000034342.13
ENSMUST00000212433.2 ENSMUST00000211809.2 |
Cklf
|
chemokine-like factor |
| chr4_-_149222057 | 0.30 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr14_-_66071412 | 0.30 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr5_-_74226394 | 0.30 |
ENSMUST00000145016.3
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr17_+_56610321 | 0.30 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr1_+_173458458 | 0.29 |
ENSMUST00000056071.13
|
Ifi209
|
interferon activated gene 209 |
| chr10_+_81012465 | 0.29 |
ENSMUST00000047864.11
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr1_+_45350698 | 0.28 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr10_-_88520877 | 0.27 |
ENSMUST00000138734.2
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr7_-_126398343 | 0.27 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr13_+_21906214 | 0.27 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr2_-_152672185 | 0.26 |
ENSMUST00000140436.2
|
Bcl2l1
|
BCL2-like 1 |
| chr12_-_113649535 | 0.26 |
ENSMUST00000103449.4
ENSMUST00000195707.3 |
Ighv2-5
|
immunoglobulin heavy variable 2-5 |
| chr13_-_97897139 | 0.26 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chr17_+_56610396 | 0.25 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr5_+_117271632 | 0.25 |
ENSMUST00000179276.8
ENSMUST00000092889.12 ENSMUST00000145640.8 |
Taok3
|
TAO kinase 3 |
| chr17_+_29709723 | 0.24 |
ENSMUST00000024811.9
|
Pim1
|
proviral integration site 1 |
| chr3_-_73615535 | 0.24 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
| chrX_-_23132991 | 0.23 |
ENSMUST00000115316.9
|
Klhl13
|
kelch-like 13 |
| chr8_-_61407760 | 0.23 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr3_+_84859453 | 0.23 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr9_+_96140750 | 0.23 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr15_-_98851566 | 0.23 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr13_-_22227114 | 0.22 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr9_+_113641615 | 0.22 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr7_-_126398165 | 0.21 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr13_-_23755374 | 0.21 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr4_+_152123772 | 0.20 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr1_+_135945798 | 0.20 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr6_-_28397997 | 0.20 |
ENSMUST00000035930.11
|
Zfp800
|
zinc finger protein 800 |
| chr2_+_120331693 | 0.19 |
ENSMUST00000141181.9
|
Capn3
|
calpain 3 |
| chr1_-_9818601 | 0.19 |
ENSMUST00000057438.7
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr13_+_22227359 | 0.19 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr3_-_102871440 | 0.19 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr4_+_11758147 | 0.19 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr7_-_35096133 | 0.19 |
ENSMUST00000154597.2
ENSMUST00000032704.12 |
Faap24
|
Fanconi anemia core complex associated protein 24 |
| chr1_-_138103021 | 0.18 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr10_+_75868432 | 0.18 |
ENSMUST00000218627.2
ENSMUST00000061617.7 |
Zfp280b
|
zinc finger protein 280B |
| chr2_+_87610895 | 0.18 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152 |
| chr19_+_12647803 | 0.18 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr12_+_76884182 | 0.18 |
ENSMUST00000041008.10
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr9_-_66951151 | 0.18 |
ENSMUST00000113696.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr3_-_73615732 | 0.18 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr12_-_65113457 | 0.18 |
ENSMUST00000221608.2
|
Fkbp3
|
FK506 binding protein 3 |
| chr11_-_69649452 | 0.17 |
ENSMUST00000058470.16
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr6_-_87798613 | 0.17 |
ENSMUST00000204169.2
|
Gm45140
|
predicted gene 45140 |
| chr12_+_98234884 | 0.17 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr17_-_27816151 | 0.17 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr3_+_135531409 | 0.17 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr12_-_115471634 | 0.17 |
ENSMUST00000103535.3
|
Ighv1-64
|
immunoglobulin heavy variable 1-64 |
| chr10_+_25317309 | 0.17 |
ENSMUST00000217929.2
ENSMUST00000220121.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr2_+_121340296 | 0.17 |
ENSMUST00000110603.2
|
Wdr76
|
WD repeat domain 76 |
| chr8_+_31640332 | 0.16 |
ENSMUST00000209851.2
ENSMUST00000098842.3 ENSMUST00000210129.2 |
Tti2
|
TELO2 interacting protein 2 |
| chr12_-_114621406 | 0.16 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr19_+_8919228 | 0.16 |
ENSMUST00000096240.3
|
Mta2
|
metastasis-associated gene family, member 2 |
| chr9_-_66951114 | 0.16 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr13_+_23935088 | 0.16 |
ENSMUST00000078369.3
|
H2ac4
|
H2A clustered histone 4 |
| chr11_-_8961109 | 0.15 |
ENSMUST00000020683.10
|
Hus1
|
HUS1 checkpoint clamp component |
| chr7_-_110443557 | 0.15 |
ENSMUST00000177462.8
ENSMUST00000176716.3 ENSMUST00000176746.8 ENSMUST00000177236.8 |
Rnf141
|
ring finger protein 141 |
| chr11_-_17903861 | 0.15 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr8_-_47070201 | 0.15 |
ENSMUST00000210264.2
ENSMUST00000040468.16 ENSMUST00000209787.2 |
Primpol
|
primase and polymerase (DNA-directed) |
| chr16_+_36755338 | 0.15 |
ENSMUST00000023531.15
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr18_+_7905440 | 0.15 |
ENSMUST00000170854.2
|
Wac
|
WW domain containing adaptor with coiled-coil |
| chr5_-_100968795 | 0.15 |
ENSMUST00000117364.8
ENSMUST00000055245.13 |
Abraxas1
|
BRCA1 A complex subunit |
| chr15_-_82223065 | 0.15 |
ENSMUST00000229733.2
ENSMUST00000229388.2 |
Naga
|
N-acetyl galactosaminidase, alpha |
| chr9_-_66951234 | 0.15 |
ENSMUST00000113690.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr6_+_67890534 | 0.15 |
ENSMUST00000197406.5
ENSMUST00000103311.3 |
Igkv11-125
|
immunoglobulin kappa variable 11-125 |
| chr2_+_112069813 | 0.14 |
ENSMUST00000028554.4
|
Lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr12_-_114443071 | 0.14 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr3_+_31150982 | 0.14 |
ENSMUST00000118204.2
|
Skil
|
SKI-like |
| chr7_-_139572452 | 0.14 |
ENSMUST00000026546.16
ENSMUST00000106069.9 ENSMUST00000148670.8 |
Adam8
|
a disintegrin and metallopeptidase domain 8 |
| chr11_+_70396070 | 0.13 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr10_-_21036792 | 0.13 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr5_-_3943907 | 0.13 |
ENSMUST00000117463.2
ENSMUST00000044746.5 |
Mterf1a
|
mitochondrial transcription termination factor 1a |
| chr7_+_65710086 | 0.13 |
ENSMUST00000153609.8
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr5_-_113970664 | 0.13 |
ENSMUST00000199109.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr6_-_68713748 | 0.13 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr1_-_118239146 | 0.13 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr10_-_28862289 | 0.13 |
ENSMUST00000152363.8
ENSMUST00000015663.7 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr6_-_37419030 | 0.13 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr3_+_30656204 | 0.13 |
ENSMUST00000192715.6
|
Mynn
|
myoneurin |
| chr2_+_85809620 | 0.13 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr2_-_37249247 | 0.13 |
ENSMUST00000112940.8
ENSMUST00000009174.15 |
Pdcl
|
phosducin-like |
| chr18_-_24153363 | 0.13 |
ENSMUST00000153337.2
ENSMUST00000148525.2 |
Zfp24
|
zinc finger protein 24 |
| chr4_-_106536063 | 0.13 |
ENSMUST00000106772.10
ENSMUST00000135676.2 ENSMUST00000026480.13 |
Ttc4
|
tetratricopeptide repeat domain 4 |
| chr3_+_103821413 | 0.13 |
ENSMUST00000051139.13
ENSMUST00000068879.11 |
Rsbn1
|
rosbin, round spermatid basic protein 1 |
| chr12_-_114579938 | 0.13 |
ENSMUST00000195469.6
ENSMUST00000109711.4 |
Ighv1-12
|
immunoglobulin heavy variable V1-12 |
| chr10_-_21036824 | 0.12 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr3_+_132335575 | 0.12 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr9_+_111011327 | 0.12 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr3_+_144824325 | 0.12 |
ENSMUST00000098538.9
ENSMUST00000106192.9 ENSMUST00000098539.7 ENSMUST00000029920.15 |
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr9_+_111011388 | 0.12 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr9_+_108820846 | 0.12 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr9_-_77255069 | 0.12 |
ENSMUST00000184848.8
ENSMUST00000184415.8 |
Mlip
|
muscular LMNA-interacting protein |
| chr3_+_30656766 | 0.12 |
ENSMUST00000047502.9
|
Mynn
|
myoneurin |
| chr12_-_114547622 | 0.12 |
ENSMUST00000193893.6
ENSMUST00000103498.3 |
Ighv1-9
|
immunoglobulin heavy variable V1-9 |
| chr1_-_138102972 | 0.12 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr13_+_23758555 | 0.12 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr18_+_37453427 | 0.11 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr3_-_92493507 | 0.11 |
ENSMUST00000194965.6
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr19_-_33764859 | 0.11 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr10_-_25076008 | 0.11 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr7_-_143102524 | 0.11 |
ENSMUST00000208093.2
ENSMUST00000209098.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr12_-_115122455 | 0.11 |
ENSMUST00000103523.2
|
Ighv1-53
|
immunoglobulin heavy variable 1-53 |
| chr2_-_110781268 | 0.11 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr12_-_115766700 | 0.11 |
ENSMUST00000196587.5
ENSMUST00000103543.3 |
Ighv1-74
|
immunoglobulin heavy variable V1-74 |
| chr8_+_47070326 | 0.11 |
ENSMUST00000211115.2
ENSMUST00000093517.7 |
Casp3
|
caspase 3 |
| chr13_+_49761506 | 0.10 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr17_+_23945310 | 0.10 |
ENSMUST00000024701.9
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr13_+_83721696 | 0.10 |
ENSMUST00000197146.5
ENSMUST00000185052.6 ENSMUST00000195984.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr6_+_11926757 | 0.10 |
ENSMUST00000133776.2
|
Phf14
|
PHD finger protein 14 |
| chr10_-_129107354 | 0.10 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr16_-_59092995 | 0.10 |
ENSMUST00000216834.2
|
Olfr201
|
olfactory receptor 201 |
| chr9_+_38164070 | 0.10 |
ENSMUST00000213129.2
|
Olfr143
|
olfactory receptor 143 |
| chr12_-_113958518 | 0.10 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr9_-_39465349 | 0.10 |
ENSMUST00000215505.2
ENSMUST00000217227.2 |
Olfr958
|
olfactory receptor 958 |
| chr4_+_98919183 | 0.10 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr6_+_129326927 | 0.10 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr17_+_21031817 | 0.09 |
ENSMUST00000232810.2
ENSMUST00000233712.2 ENSMUST00000232852.2 |
Vmn1r229
|
vomeronasal 1 receptor 229 |
| chr2_+_29236815 | 0.09 |
ENSMUST00000028139.11
ENSMUST00000113830.11 |
Med27
|
mediator complex subunit 27 |
| chr7_-_28071919 | 0.09 |
ENSMUST00000119990.8
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr7_-_126391657 | 0.09 |
ENSMUST00000032936.8
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr5_-_5315968 | 0.09 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr12_-_114023935 | 0.09 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr9_-_19275301 | 0.09 |
ENSMUST00000214810.2
|
Olfr846
|
olfactory receptor 846 |
| chr15_+_6451721 | 0.09 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr4_+_19280850 | 0.09 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr3_+_135531834 | 0.09 |
ENSMUST00000029810.6
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr11_-_101917745 | 0.09 |
ENSMUST00000107167.2
ENSMUST00000062801.11 |
Mpp3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr1_-_4479445 | 0.09 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr12_-_115083839 | 0.09 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr7_-_84661476 | 0.09 |
ENSMUST00000124773.3
ENSMUST00000233725.2 ENSMUST00000233739.2 ENSMUST00000232837.2 |
Vmn2r66
|
vomeronasal 2, receptor 66 |
| chr10_-_80257681 | 0.09 |
ENSMUST00000156244.2
|
Tcf3
|
transcription factor 3 |
| chr19_-_46315543 | 0.09 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr2_-_37249208 | 0.09 |
ENSMUST00000147703.3
|
Pdcl
|
phosducin-like |
| chr15_+_41694317 | 0.08 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr12_-_114502585 | 0.08 |
ENSMUST00000103496.4
|
Ighv1-7
|
immunoglobulin heavy variable V1-7 |
| chr10_+_94412116 | 0.08 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr15_-_42540363 | 0.08 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr10_+_5543769 | 0.08 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr7_-_126391388 | 0.08 |
ENSMUST00000206570.2
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr9_-_77255099 | 0.08 |
ENSMUST00000184138.8
ENSMUST00000184006.8 ENSMUST00000185144.8 ENSMUST00000034910.16 |
Mlip
|
muscular LMNA-interacting protein |
| chr11_+_57409484 | 0.08 |
ENSMUST00000108849.8
ENSMUST00000020830.14 |
Mfap3
|
microfibrillar-associated protein 3 |
| chr8_+_12999480 | 0.08 |
ENSMUST00000110866.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr17_+_64170045 | 0.08 |
ENSMUST00000233225.2
|
Fer
|
fer (fms/fps related) protein kinase |
| chr9_-_39863342 | 0.08 |
ENSMUST00000216647.2
|
Olfr975
|
olfactory receptor 975 |
| chr12_-_115172211 | 0.08 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr6_+_57234937 | 0.08 |
ENSMUST00000228297.2
|
Vmn1r15
|
vomeronasal 1 receptor 15 |
| chr1_+_143614753 | 0.08 |
ENSMUST00000145969.8
|
Glrx2
|
glutaredoxin 2 (thioltransferase) |
| chr17_-_80203457 | 0.08 |
ENSMUST00000068282.7
ENSMUST00000112437.8 |
Atl2
|
atlastin GTPase 2 |
| chr1_+_87331427 | 0.07 |
ENSMUST00000172736.8
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr6_-_69877642 | 0.07 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr9_+_3000922 | 0.07 |
ENSMUST00000151376.3
|
Gm10722
|
predicted gene 10722 |
| chr2_+_85780781 | 0.07 |
ENSMUST00000080698.3
|
Olfr1028
|
olfactory receptor 1028 |
| chr10_-_25172953 | 0.07 |
ENSMUST00000177124.2
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr7_+_107679062 | 0.07 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.2 | 1.0 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.9 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 1.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.3 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.2 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.2 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.1 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.0 | 0.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.7 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.5 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.0 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.5 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.2 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 2.9 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.0 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0043056 | forward locomotion(GO:0043056) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.2 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) dense core granule membrane(GO:0032127) |
| 0.0 | 0.8 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0099738 | cell cortex region(GO:0099738) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:0050656 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.3 | 1.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.4 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.1 | 0.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008456 | alpha-N-acetylgalactosaminidase activity(GO:0008456) |
| 0.0 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 2.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 0.0 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 6.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0004854 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.6 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |