Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pou2f2_Pou3f1
Z-value: 1.59
Transcription factors associated with Pou2f2_Pou3f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
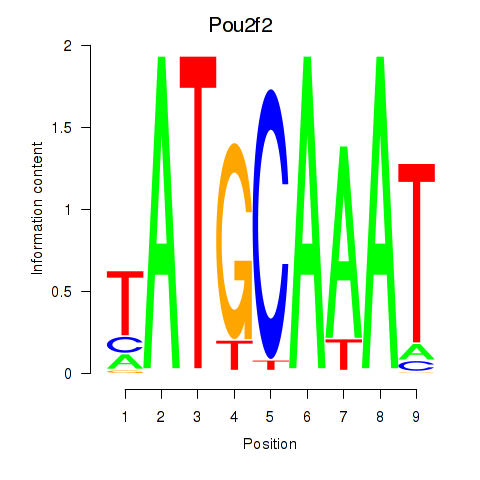
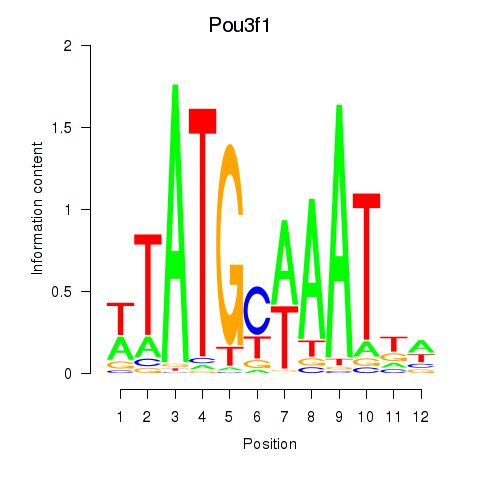
Pou2f2
|
ENSMUSG00000008496.20 | POU domain, class 2, transcription factor 2 |
|
Pou3f1
|
ENSMUSG00000090125.4 | POU domain, class 3, transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f2 | mm39_v1_chr7_-_24831892_24831937 | 0.53 | 8.5e-04 | Click! |
| Pou3f1 | mm39_v1_chr4_+_124550600_124550600 | -0.17 | 3.2e-01 | Click! |
Activity profile of Pou2f2_Pou3f1 motif
Sorted Z-values of Pou2f2_Pou3f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_67014383 | 13.29 |
ENSMUST00000043098.9
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr11_-_69496655 | 12.78 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr11_+_95227836 | 10.54 |
ENSMUST00000037502.7
|
Fam117a
|
family with sequence similarity 117, member A |
| chr7_-_4815542 | 10.48 |
ENSMUST00000079496.9
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr12_-_36092475 | 8.22 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr15_-_66703471 | 8.12 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr11_+_58839716 | 7.76 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chr6_-_67014348 | 7.63 |
ENSMUST00000204369.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr10_+_75784126 | 7.48 |
ENSMUST00000000926.3
|
Vpreb3
|
pre-B lymphocyte gene 3 |
| chr8_+_23464860 | 7.24 |
ENSMUST00000110688.9
ENSMUST00000121802.9 |
Ank1
|
ankyrin 1, erythroid |
| chr6_-_68713748 | 7.11 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr1_+_91468409 | 7.00 |
ENSMUST00000027538.9
ENSMUST00000190484.7 ENSMUST00000186068.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr6_+_145091196 | 6.79 |
ENSMUST00000156849.8
ENSMUST00000132948.2 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr17_+_29709723 | 6.74 |
ENSMUST00000024811.9
|
Pim1
|
proviral integration site 1 |
| chr3_+_51568588 | 6.57 |
ENSMUST00000099106.10
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr6_-_67014191 | 6.40 |
ENSMUST00000204282.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr13_+_23930717 | 6.23 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chrX_+_92718695 | 6.09 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr11_-_106205320 | 5.82 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chrX_+_55500170 | 5.54 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr13_-_22227114 | 5.44 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr17_+_27775637 | 5.27 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr5_+_137627431 | 5.25 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr17_+_27775613 | 5.24 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_27775471 | 5.17 |
ENSMUST00000118599.9
ENSMUST00000232265.2 ENSMUST00000232013.2 ENSMUST00000114888.11 ENSMUST00000231874.2 ENSMUST00000119486.9 ENSMUST00000231825.2 ENSMUST00000231866.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr13_+_22227359 | 4.86 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr7_+_126376099 | 4.76 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr5_+_137628377 | 4.68 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr12_-_113542610 | 4.66 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr1_+_91468266 | 4.52 |
ENSMUST00000086843.11
|
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr14_-_67953035 | 4.52 |
ENSMUST00000163100.8
ENSMUST00000132705.8 ENSMUST00000124045.3 |
Cdca2
|
cell division cycle associated 2 |
| chr4_-_152561896 | 4.51 |
ENSMUST00000238738.2
ENSMUST00000162017.3 ENSMUST00000030768.10 |
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr12_-_113823290 | 4.51 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr11_+_58845502 | 4.44 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr4_+_152123772 | 4.42 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chrX_-_165992311 | 4.38 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr12_-_113561594 | 4.36 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chrX_-_165992145 | 4.35 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr10_-_88520877 | 4.21 |
ENSMUST00000138734.2
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr12_-_113589576 | 4.16 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr6_-_56878854 | 3.99 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr12_-_114443071 | 3.92 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr7_+_126376353 | 3.92 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr13_-_23755374 | 3.75 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr19_-_46033353 | 3.73 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr12_-_113802603 | 3.71 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr13_+_21906214 | 3.68 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr19_+_8919228 | 3.64 |
ENSMUST00000096240.3
|
Mta2
|
metastasis-associated gene family, member 2 |
| chr13_-_22017677 | 3.60 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr7_-_143014726 | 3.57 |
ENSMUST00000167912.9
ENSMUST00000037287.8 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C (P57) |
| chr6_-_69800923 | 3.56 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr3_+_96177010 | 3.47 |
ENSMUST00000051089.4
ENSMUST00000177113.2 |
Gm42743
H2bc18
|
predicted gene 42743 H2B clustered histone 18 |
| chr6_+_67586695 | 3.46 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr13_+_23935088 | 3.39 |
ENSMUST00000078369.3
|
H2ac4
|
H2A clustered histone 4 |
| chr17_-_35383867 | 3.30 |
ENSMUST00000025253.12
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr7_+_44667377 | 3.25 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr2_-_152239966 | 3.18 |
ENSMUST00000063332.9
ENSMUST00000182625.2 |
Sox12
|
SRY (sex determining region Y)-box 12 |
| chr11_-_5787743 | 3.18 |
ENSMUST00000109837.8
|
Polm
|
polymerase (DNA directed), mu |
| chr12_-_113666198 | 3.16 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr12_-_113860566 | 3.14 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr2_+_160487801 | 3.01 |
ENSMUST00000109468.3
|
Top1
|
topoisomerase (DNA) I |
| chr12_-_114752425 | 2.99 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr16_+_91526169 | 2.98 |
ENSMUST00000114001.8
ENSMUST00000113999.8 ENSMUST00000064797.12 ENSMUST00000114002.9 ENSMUST00000095909.10 ENSMUST00000056482.14 ENSMUST00000113996.8 |
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr6_-_68609426 | 2.98 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr10_+_79762858 | 2.87 |
ENSMUST00000019708.12
ENSMUST00000105377.8 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr13_+_21994588 | 2.81 |
ENSMUST00000091745.6
|
H2ac23
|
H2A clustered histone 23 |
| chr12_-_114355789 | 2.81 |
ENSMUST00000103486.2
|
Ighv6-3
|
immunoglobulin heavy variable 6-3 |
| chr7_-_44180700 | 2.75 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr14_+_26722319 | 2.74 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr9_+_71123061 | 2.71 |
ENSMUST00000034723.6
|
Aldh1a2
|
aldehyde dehydrogenase family 1, subfamily A2 |
| chr2_+_25070749 | 2.68 |
ENSMUST00000104999.4
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr6_+_68247469 | 2.63 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr10_-_80257681 | 2.62 |
ENSMUST00000156244.2
|
Tcf3
|
transcription factor 3 |
| chr13_+_22220000 | 2.57 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr12_-_114487525 | 2.56 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr8_+_71151581 | 2.55 |
ENSMUST00000095267.8
|
Jund
|
jun D proto-oncogene |
| chr4_-_128699838 | 2.52 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr12_-_114321838 | 2.44 |
ENSMUST00000125484.3
|
Ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr8_-_65489834 | 2.42 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr6_+_68279392 | 2.39 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr8_+_121215155 | 2.37 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr9_-_22300409 | 2.33 |
ENSMUST00000040912.9
|
Anln
|
anillin, actin binding protein |
| chr6_-_69741999 | 2.33 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr19_+_47217279 | 2.31 |
ENSMUST00000111807.5
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr12_-_113912416 | 2.29 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr11_+_98632631 | 2.28 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr2_+_90927053 | 2.27 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr12_-_113790741 | 2.27 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr2_-_152672535 | 2.26 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr8_+_71069476 | 2.25 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr6_+_68026941 | 2.23 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr6_+_136495784 | 2.20 |
ENSMUST00000032335.13
ENSMUST00000185724.7 |
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr2_-_102282844 | 2.20 |
ENSMUST00000099678.5
|
Fjx1
|
four jointed box 1 |
| chrX_+_132751729 | 2.19 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr8_-_123187406 | 2.00 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr7_+_24981604 | 1.99 |
ENSMUST00000163320.8
ENSMUST00000005578.13 |
Cic
|
capicua transcriptional repressor |
| chr2_-_152672185 | 1.98 |
ENSMUST00000140436.2
|
Bcl2l1
|
BCL2-like 1 |
| chr7_+_127345909 | 1.94 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr12_-_113700190 | 1.94 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr7_+_27186335 | 1.92 |
ENSMUST00000008528.8
|
Sertad1
|
SERTA domain containing 1 |
| chr6_+_68098030 | 1.91 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr12_-_114502585 | 1.86 |
ENSMUST00000103496.4
|
Ighv1-7
|
immunoglobulin heavy variable V1-7 |
| chr12_-_114226570 | 1.85 |
ENSMUST00000103479.4
ENSMUST00000195619.2 |
Ighv3-5
|
immunoglobulin heavy variable 3-5 |
| chr7_+_121888520 | 1.85 |
ENSMUST00000064989.12
ENSMUST00000064921.5 |
Prkcb
|
protein kinase C, beta |
| chr6_-_69792108 | 1.83 |
ENSMUST00000103367.3
|
Igkv12-44
|
immunoglobulin kappa variable 12-44 |
| chr4_-_63965161 | 1.81 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr8_-_65489791 | 1.79 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr13_+_83723255 | 1.76 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr7_+_24206431 | 1.72 |
ENSMUST00000176880.2
|
Zfp428
|
zinc finger protein 428 |
| chr13_-_22219738 | 1.70 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr6_+_67890534 | 1.66 |
ENSMUST00000197406.5
ENSMUST00000103311.3 |
Igkv11-125
|
immunoglobulin kappa variable 11-125 |
| chr7_+_24206482 | 1.64 |
ENSMUST00000071361.13
|
Zfp428
|
zinc finger protein 428 |
| chr19_+_53128901 | 1.62 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr10_-_79911245 | 1.59 |
ENSMUST00000217972.2
|
Sbno2
|
strawberry notch 2 |
| chr13_+_23718038 | 1.59 |
ENSMUST00000073261.3
|
H2ac10
|
H2A clustered histone 10 |
| chr5_+_76988444 | 1.56 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr16_+_17051423 | 1.56 |
ENSMUST00000090190.14
ENSMUST00000232082.2 ENSMUST00000232426.2 |
Hic2
Gm49573
|
hypermethylated in cancer 2 predicted gene, 49573 |
| chr12_-_114398864 | 1.56 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr13_-_95661726 | 1.55 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr12_-_114117264 | 1.55 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr3_+_83673606 | 1.53 |
ENSMUST00000029625.8
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr12_-_114252202 | 1.52 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr8_+_31640332 | 1.50 |
ENSMUST00000209851.2
ENSMUST00000098842.3 ENSMUST00000210129.2 |
Tti2
|
TELO2 interacting protein 2 |
| chr10_+_79763164 | 1.49 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr17_-_46867083 | 1.49 |
ENSMUST00000015749.7
|
Srf
|
serum response factor |
| chr12_-_114710326 | 1.48 |
ENSMUST00000103507.2
|
Ighv1-22
|
immunoglobulin heavy variable 1-22 |
| chr9_+_21848282 | 1.47 |
ENSMUST00000046371.13
|
Plppr2
|
phospholipid phosphatase related 2 |
| chr11_+_98632696 | 1.42 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr12_-_115766700 | 1.42 |
ENSMUST00000196587.5
ENSMUST00000103543.3 |
Ighv1-74
|
immunoglobulin heavy variable V1-74 |
| chr6_-_69753317 | 1.40 |
ENSMUST00000103366.3
|
Igkv5-45
|
immunoglobulin kappa chain variable 5-45 |
| chr6_-_69704122 | 1.36 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr12_-_114621406 | 1.33 |
ENSMUST00000192077.2
|
Ighv1-15
|
immunoglobulin heavy variable 1-15 |
| chr14_+_79753055 | 1.29 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr6_-_67919524 | 1.26 |
ENSMUST00000196768.2
|
Igkv9-124
|
immunoglobulin kappa chain variable 9-124 |
| chr13_+_23758555 | 1.23 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr6_-_69877961 | 1.22 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr1_+_12788720 | 1.22 |
ENSMUST00000088585.10
|
Sulf1
|
sulfatase 1 |
| chr12_-_114286421 | 1.21 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr19_+_53128861 | 1.21 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr15_+_102381705 | 1.17 |
ENSMUST00000229958.2
ENSMUST00000229184.2 ENSMUST00000230728.2 ENSMUST00000230114.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr5_+_144127102 | 1.16 |
ENSMUST00000060747.8
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr6_-_87510200 | 1.15 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr13_-_23867924 | 1.14 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chr1_+_75213082 | 1.13 |
ENSMUST00000055223.14
ENSMUST00000082158.13 ENSMUST00000188346.7 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr3_+_129974531 | 1.13 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr9_-_85209162 | 1.13 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr12_-_79239022 | 1.12 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr5_-_124003553 | 1.10 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr9_-_85209340 | 1.10 |
ENSMUST00000187711.2
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr6_+_68657317 | 1.10 |
ENSMUST00000198735.2
|
Igkv10-95
|
immunoglobulin kappa variable 10-95 |
| chr4_+_128582519 | 1.10 |
ENSMUST00000106080.8
|
Phc2
|
polyhomeotic 2 |
| chr13_+_16186410 | 1.10 |
ENSMUST00000042603.14
|
Inhba
|
inhibin beta-A |
| chr2_-_71198091 | 1.09 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr7_+_24206547 | 1.09 |
ENSMUST00000177205.2
|
Zfp428
|
zinc finger protein 428 |
| chr6_+_68402550 | 1.08 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr12_-_113958518 | 1.07 |
ENSMUST00000103467.2
|
Ighv14-2
|
immunoglobulin heavy variable 14-2 |
| chr17_-_59298338 | 1.06 |
ENSMUST00000025064.14
|
Pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr6_-_68857658 | 1.06 |
ENSMUST00000198756.2
|
Gm42543
|
predicted gene 42543 |
| chr16_-_91525485 | 1.05 |
ENSMUST00000231499.2
ENSMUST00000141664.9 ENSMUST00000123751.2 ENSMUST00000122254.8 ENSMUST00000114023.3 |
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr6_-_65121892 | 1.03 |
ENSMUST00000031982.5
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr6_-_69877642 | 1.02 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr19_+_47926086 | 1.00 |
ENSMUST00000238163.2
ENSMUST00000066308.9 |
Cfap58
|
cilia and flagella associated protein 58 |
| chrX_+_149377416 | 0.99 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr9_-_119812042 | 0.98 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr12_-_114023935 | 0.98 |
ENSMUST00000103469.4
|
Ighv14-3
|
immunoglobulin heavy variable V14-3 |
| chr13_+_37529184 | 0.95 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr10_-_79744726 | 0.95 |
ENSMUST00000165684.8
ENSMUST00000164705.8 ENSMUST00000105378.9 ENSMUST00000170409.2 |
Med16
|
mediator complex subunit 16 |
| chr1_+_75213044 | 0.92 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr6_-_69835868 | 0.91 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr5_-_123126550 | 0.91 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr5_-_123038329 | 0.91 |
ENSMUST00000031435.14
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr12_-_115172211 | 0.91 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr7_-_44702269 | 0.90 |
ENSMUST00000057293.8
|
Prr12
|
proline rich 12 |
| chr16_-_4698148 | 0.89 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr11_-_34724458 | 0.88 |
ENSMUST00000093191.3
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr2_+_118877594 | 0.88 |
ENSMUST00000152380.8
ENSMUST00000099542.9 |
Knl1
|
kinetochore scaffold 1 |
| chr11_+_83741689 | 0.88 |
ENSMUST00000108114.9
|
Hnf1b
|
HNF1 homeobox B |
| chr4_+_3678108 | 0.87 |
ENSMUST00000041377.13
ENSMUST00000103010.4 |
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr12_-_114672701 | 0.86 |
ENSMUST00000103505.3
ENSMUST00000193855.2 |
Ighv1-19
|
immunoglobulin heavy variable V1-19 |
| chr6_-_148847854 | 0.86 |
ENSMUST00000139355.8
ENSMUST00000146457.2 ENSMUST00000054080.15 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr1_+_75213114 | 0.85 |
ENSMUST00000188290.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr18_+_50184769 | 0.84 |
ENSMUST00000134348.8
ENSMUST00000153873.3 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr6_+_70495224 | 0.83 |
ENSMUST00000103396.2
|
Igkv3-12
|
immunoglobulin kappa variable 3-12 |
| chr3_+_159201048 | 0.83 |
ENSMUST00000120272.8
|
Depdc1a
|
DEP domain containing 1a |
| chr8_+_70686836 | 0.82 |
ENSMUST00000164403.8
ENSMUST00000093458.11 |
Sugp2
|
SURP and G patch domain containing 2 |
| chr12_-_114012399 | 0.82 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr3_+_96128427 | 0.81 |
ENSMUST00000090781.8
|
H2bc21
|
H2B clustered histone 21 |
| chr3_-_97134680 | 0.80 |
ENSMUST00000046521.14
|
Bcl9
|
B cell CLL/lymphoma 9 |
| chr11_+_98632953 | 0.79 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr4_-_129590609 | 0.79 |
ENSMUST00000102588.10
|
Tmem39b
|
transmembrane protein 39b |
| chr6_-_69940857 | 0.76 |
ENSMUST00000103372.3
|
Igkv5-37
|
immunoglobulin kappa variable 5-37 |
| chr10_+_128542120 | 0.76 |
ENSMUST00000054125.9
|
Pmel
|
premelanosome protein |
| chr19_-_40982576 | 0.76 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr6_+_41098273 | 0.76 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr12_-_115157739 | 0.74 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr8_-_70687051 | 0.73 |
ENSMUST00000019679.12
|
Armc6
|
armadillo repeat containing 6 |
| chr9_+_106099797 | 0.71 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr6_+_67816777 | 0.70 |
ENSMUST00000200578.5
ENSMUST00000103308.3 |
Igkv9-129
|
immunoglobulin kappa variable 9-129 |
| chrX_+_158155171 | 0.70 |
ENSMUST00000087143.7
|
Eif1ax
|
eukaryotic translation initiation factor 1A, X-linked |
| chr15_-_93417380 | 0.69 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f2_Pou3f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 12.8 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 2.1 | 10.5 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.9 | 26.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.5 | 4.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 1.2 | 3.7 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 1.1 | 4.5 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.1 | 4.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.8 | 2.3 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.8 | 1.5 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.7 | 7.5 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.7 | 1.5 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) |
| 0.7 | 7.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.6 | 1.8 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.6 | 2.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.6 | 10.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.6 | 6.7 | GO:1902033 | vitamin D receptor signaling pathway(GO:0070561) regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.5 | 8.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.5 | 1.5 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 69.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.5 | 2.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 | 2.9 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.4 | 1.6 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.4 | 1.6 | GO:0035565 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.4 | 2.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.4 | 0.7 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.3 | 3.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 2.6 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.3 | 3.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 3.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 0.9 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.3 | 15.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.3 | 1.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 2.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 0.8 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 6.6 | GO:0019370 | glutathione biosynthetic process(GO:0006750) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 4.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.7 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.2 | 1.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 1.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 0.6 | GO:1904268 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.2 | 1.8 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.2 | 1.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 2.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 4.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 0.6 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.2 | 1.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 0.5 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.2 | 2.7 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 7.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 1.8 | GO:2000111 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 1.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.1 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.1 | 0.6 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 | 28.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.8 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 0.2 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:1905066 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) blood vessel lumenization(GO:0072554) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 5.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 2.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.8 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 8.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 2.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 6.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 2.4 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 0.2 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 2.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.6 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.1 | 4.5 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 4.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.2 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 3.6 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.1 | 4.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 3.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 0.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 1.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 1.3 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 7.1 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.2 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.5 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 1.1 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 2.8 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 1.9 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 15.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.5 | 4.5 | GO:1990031 | pinceau fiber(GO:1990031) |
| 1.3 | 12.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 75.0 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.5 | 2.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.4 | 3.0 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 1.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.3 | 1.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 10.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 4.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 2.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 2.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 0.9 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 5.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.7 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 0.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 2.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 9.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 29.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 9.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 2.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 3.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 2.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 4.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 2.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 6.3 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 15.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.6 | 6.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.5 | 6.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.1 | 4.5 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.1 | 12.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.0 | 3.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.7 | 7.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.6 | 4.4 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.6 | 2.3 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.5 | 69.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.5 | 6.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 4.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 1.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 2.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 1.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 3.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 0.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 5.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 10.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 1.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 3.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 8.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.2 | 4.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 2.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 2.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 30.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 0.9 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 2.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 2.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 7.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 5.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 3.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 3.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 1.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.3 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 1.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 3.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 3.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 3.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.1 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 2.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 27.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 20.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 7.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 6.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 4.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 9.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 3.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 15.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 12.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 4.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 6.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 4.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.2 | 3.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 8.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 6.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 5.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 10.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 21.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 3.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 3.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |