Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
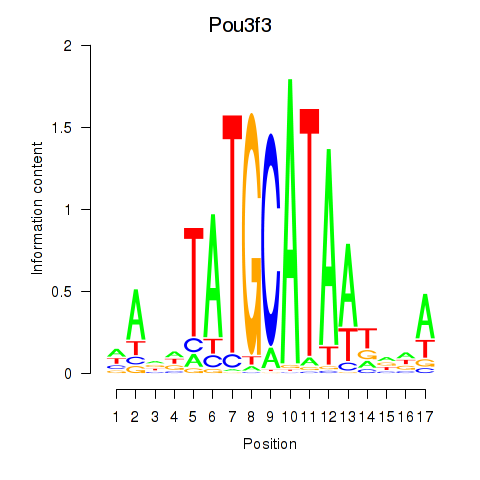
Results for Pou3f3
Z-value: 0.49
Transcription factors associated with Pou3f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f3
|
ENSMUSG00000045515.5 | POU domain, class 3, transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f3 | mm39_v1_chr1_+_42734051_42734105 | 0.10 | 5.5e-01 | Click! |
Activity profile of Pou3f3 motif
Sorted Z-values of Pou3f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_134238310 | 4.31 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr4_-_149221998 | 2.95 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr5_+_66833434 | 2.88 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr4_-_149222057 | 2.50 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr6_+_129326927 | 2.12 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr14_+_26722319 | 2.02 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr5_-_74226394 | 1.76 |
ENSMUST00000145016.3
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr7_-_126391657 | 1.32 |
ENSMUST00000032936.8
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr7_-_126391388 | 1.31 |
ENSMUST00000206570.2
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr13_+_21906214 | 1.12 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr8_-_106022676 | 0.81 |
ENSMUST00000057855.4
|
Exoc3l
|
exocyst complex component 3-like |
| chr11_-_99134885 | 0.76 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr15_-_42540363 | 0.68 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr3_+_83673606 | 0.68 |
ENSMUST00000029625.8
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr1_+_43571536 | 0.55 |
ENSMUST00000202540.2
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr3_-_122828592 | 0.50 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr12_-_113912416 | 0.34 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr3_-_144555062 | 0.28 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr2_-_154214622 | 0.24 |
ENSMUST00000028990.10
ENSMUST00000109730.3 |
Cdk5rap1
|
CDK5 regulatory subunit associated protein 1 |
| chr2_+_164664920 | 0.13 |
ENSMUST00000132282.2
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr2_+_67004178 | 0.11 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr6_+_96092230 | 0.07 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr10_+_26105605 | 0.05 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr3_+_92223927 | 0.04 |
ENSMUST00000061038.4
|
Sprr2b
|
small proline-rich protein 2B |
| chr3_-_133250889 | 0.03 |
ENSMUST00000197118.5
|
Tet2
|
tet methylcytosine dioxygenase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 5.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 0.7 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 | 2.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 0.7 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.5 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 2.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 4.3 | GO:0007051 | spindle organization(GO:0007051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.4 | 2.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.3 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 2.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 1.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |