Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pou4f3
Z-value: 0.36
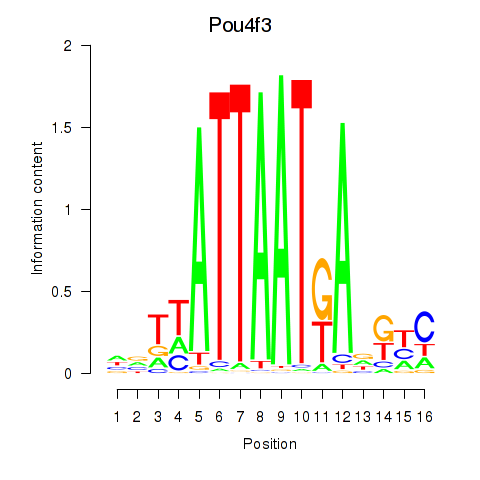
Transcription factors associated with Pou4f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f3
|
ENSMUSG00000024497.5 | POU domain, class 4, transcription factor 3 |
Activity profile of Pou4f3 motif
Sorted Z-values of Pou4f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41498716 | 2.80 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr4_-_119047202 | 0.87 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr8_-_84184978 | 0.84 |
ENSMUST00000081506.11
|
Scoc
|
short coiled-coil protein |
| chr4_-_119047167 | 0.70 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047180 | 0.54 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr10_-_37014859 | 0.52 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr4_-_119047146 | 0.52 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr18_+_11766333 | 0.48 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr4_-_108158242 | 0.44 |
ENSMUST00000043616.7
|
Zyg11b
|
zyg-ll family member B, cell cycle regulator |
| chr1_+_45834645 | 0.41 |
ENSMUST00000147308.2
|
Wdr75
|
WD repeat domain 75 |
| chr1_-_36982747 | 0.37 |
ENSMUST00000185964.3
|
Tmem131
|
transmembrane protein 131 |
| chr2_+_36120438 | 0.35 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr1_+_107456731 | 0.34 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr14_-_75991903 | 0.33 |
ENSMUST00000049168.9
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr9_+_109961079 | 0.33 |
ENSMUST00000197480.5
ENSMUST00000197984.5 ENSMUST00000199896.2 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr10_+_97318223 | 0.31 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr7_-_37718916 | 0.26 |
ENSMUST00000085513.6
ENSMUST00000206327.2 |
Uri1
|
URI1, prefoldin-like chaperone |
| chr13_+_24023386 | 0.26 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr9_+_109961048 | 0.25 |
ENSMUST00000088716.12
|
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr14_+_26722319 | 0.24 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr19_+_29902506 | 0.24 |
ENSMUST00000120388.9
ENSMUST00000144528.8 ENSMUST00000177518.8 |
Il33
|
interleukin 33 |
| chr13_+_24023428 | 0.23 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr12_-_25147139 | 0.21 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr9_+_111011388 | 0.21 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr1_+_179788675 | 0.20 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr9_-_15212745 | 0.20 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chrY_-_1245685 | 0.19 |
ENSMUST00000143286.8
ENSMUST00000137048.8 ENSMUST00000069309.14 ENSMUST00000139365.8 ENSMUST00000154004.8 |
Uty
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr2_+_3425159 | 0.19 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr18_+_56695515 | 0.18 |
ENSMUST00000130163.8
ENSMUST00000132628.8 |
Phax
|
phosphorylated adaptor for RNA export |
| chr17_-_33252341 | 0.18 |
ENSMUST00000087654.5
|
Zfp763
|
zinc finger protein 763 |
| chr9_+_111011327 | 0.18 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_+_132689640 | 0.16 |
ENSMUST00000124836.8
ENSMUST00000154160.2 |
Crls1
|
cardiolipin synthase 1 |
| chr4_+_102446883 | 0.14 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr3_-_86827664 | 0.13 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr1_-_126758369 | 0.13 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chrX_-_74423647 | 0.12 |
ENSMUST00000114085.9
|
F8
|
coagulation factor VIII |
| chr5_-_69748126 | 0.12 |
ENSMUST00000166298.8
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chrX_+_70860378 | 0.11 |
ENSMUST00000114575.4
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr10_-_75946790 | 0.10 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr17_-_50401305 | 0.10 |
ENSMUST00000113195.8
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr3_-_86827640 | 0.09 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr12_-_113802603 | 0.09 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr5_+_87148697 | 0.09 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr1_+_179788037 | 0.08 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr18_+_23886765 | 0.08 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr16_+_11224481 | 0.08 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29 |
| chr6_-_138020409 | 0.07 |
ENSMUST00000111873.8
ENSMUST00000141280.3 |
Slc15a5
|
solute carrier family 15, member 5 |
| chr14_+_69585036 | 0.07 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr15_+_79575046 | 0.06 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr1_+_139429430 | 0.05 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr18_-_68562385 | 0.05 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr9_-_15212849 | 0.05 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr9_-_99302205 | 0.04 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chrX_+_9751861 | 0.04 |
ENSMUST00000067529.9
ENSMUST00000086165.4 |
Sytl5
|
synaptotagmin-like 5 |
| chr11_+_109434519 | 0.04 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr2_+_4305273 | 0.04 |
ENSMUST00000175669.8
|
Frmd4a
|
FERM domain containing 4A |
| chr4_-_43710231 | 0.03 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr4_+_63477018 | 0.03 |
ENSMUST00000077709.11
|
Tmem268
|
transmembrane protein 268 |
| chrX_+_70860357 | 0.02 |
ENSMUST00000114576.9
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr7_-_99629637 | 0.02 |
ENSMUST00000080817.6
|
Rnf169
|
ring finger protein 169 |
| chr5_-_38649291 | 0.02 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr9_+_109881083 | 0.01 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chr3_-_72875187 | 0.01 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr10_-_129107354 | 0.01 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777 |
| chr15_+_66542598 | 0.01 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin |
| chr5_+_13448833 | 0.01 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr8_+_15107646 | 0.00 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou4f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.4 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.4 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |