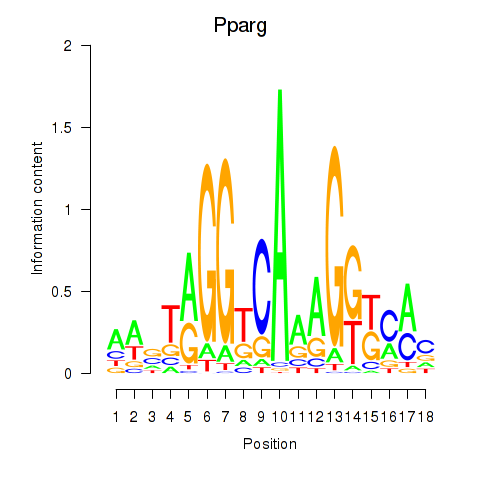
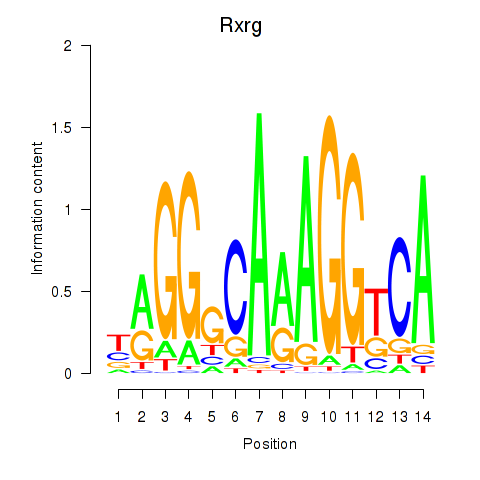
Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Pparg_Rxrg
Z-value: 4.54
Transcription factors associated with Pparg_Rxrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pparg
|
ENSMUSG00000000440.13 | peroxisome proliferator activated receptor gamma |
|
Rxrg
|
ENSMUSG00000015843.11 | retinoid X receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrg | mm39_v1_chr1_+_167445815_167445878 | 0.92 | 1.1e-15 | Click! |
| Pparg | mm39_v1_chr6_+_115398996_115399030 | 0.47 | 3.8e-03 | Click! |
Activity profile of Pparg_Rxrg motif
Sorted Z-values of Pparg_Rxrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_46120327 | 110.55 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr19_+_39275518 | 109.62 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr17_-_46749370 | 86.39 |
ENSMUST00000087012.7
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr9_-_46146558 | 81.67 |
ENSMUST00000121916.8
ENSMUST00000034586.9 |
Apoc3
|
apolipoprotein C-III |
| chr7_+_26821266 | 71.67 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr9_-_46146928 | 71.48 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr15_+_82336535 | 65.71 |
ENSMUST00000089129.7
ENSMUST00000229313.2 ENSMUST00000231136.2 |
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr15_-_82648376 | 56.91 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr3_+_138121245 | 46.20 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr9_-_15212849 | 43.10 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr2_+_172994841 | 41.33 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr17_-_46749320 | 39.41 |
ENSMUST00000233575.2
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr11_+_69945157 | 38.23 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr10_+_127637015 | 37.99 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr19_-_8382424 | 35.72 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr4_-_62005498 | 34.36 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr4_-_61700450 | 32.69 |
ENSMUST00000107477.2
ENSMUST00000080606.9 |
Mup19
|
major urinary protein 19 |
| chr3_+_94600863 | 32.22 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr19_-_39637489 | 30.13 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr9_-_86577940 | 29.11 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr7_-_99345016 | 28.69 |
ENSMUST00000107086.9
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr19_-_40175709 | 28.20 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr7_-_97066937 | 27.46 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr15_+_76579960 | 25.75 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr19_+_12610668 | 25.17 |
ENSMUST00000044976.12
|
Glyat
|
glycine-N-acyltransferase |
| chr5_-_110434026 | 24.26 |
ENSMUST00000031472.12
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chr7_+_140343652 | 24.06 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr6_-_85846110 | 23.88 |
ENSMUST00000045008.8
|
Nat8f2
|
N-acetyltransferase 8 (GCN5-related) family member 2 |
| chr2_-_25390625 | 23.57 |
ENSMUST00000040042.11
|
C8g
|
complement component 8, gamma polypeptide |
| chr15_+_82439273 | 23.16 |
ENSMUST00000229103.2
ENSMUST00000068861.8 ENSMUST00000229904.2 |
Cyp2d12
|
cytochrome P450, family 2, subfamily d, polypeptide 12 |
| chr5_+_114284585 | 22.82 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr5_-_130053120 | 22.65 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr11_-_77784922 | 22.36 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr17_-_35100980 | 22.27 |
ENSMUST00000152417.8
ENSMUST00000146299.8 |
C2
Gm20547
|
complement component 2 (within H-2S) predicted gene 20547 |
| chr7_-_99344779 | 21.62 |
ENSMUST00000137914.2
ENSMUST00000207090.2 ENSMUST00000208225.2 |
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr15_+_76579885 | 21.60 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr13_-_42000958 | 21.53 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr17_-_74257164 | 21.38 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr13_-_93774469 | 21.01 |
ENSMUST00000099309.6
|
Bhmt
|
betaine-homocysteine methyltransferase |
| chr10_-_128796834 | 20.86 |
ENSMUST00000026398.5
|
Mettl7b
|
methyltransferase like 7B |
| chr7_+_140415431 | 20.63 |
ENSMUST00000209978.2
ENSMUST00000210916.2 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr9_-_15212745 | 20.39 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr17_-_35101069 | 20.15 |
ENSMUST00000025230.15
|
C2
|
complement component 2 (within H-2S) |
| chr19_+_39980868 | 20.10 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr4_-_60697274 | 20.04 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr6_-_85797946 | 19.92 |
ENSMUST00000032074.5
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr2_-_32594156 | 19.79 |
ENSMUST00000127812.3
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr9_+_46180362 | 19.74 |
ENSMUST00000214202.2
ENSMUST00000215458.2 ENSMUST00000215187.2 ENSMUST00000213878.2 ENSMUST00000034584.4 |
Apoa5
|
apolipoprotein A-V |
| chr1_-_180023518 | 19.70 |
ENSMUST00000162769.8
ENSMUST00000161379.2 ENSMUST00000027766.13 ENSMUST00000161814.8 |
Coq8a
|
coenzyme Q8A |
| chr9_+_46151994 | 19.49 |
ENSMUST00000034585.7
|
Apoa4
|
apolipoprotein A-IV |
| chr10_+_87694924 | 18.95 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr11_+_75358866 | 18.92 |
ENSMUST00000043598.14
ENSMUST00000108435.2 |
Tlcd2
|
TLC domain containing 2 |
| chr7_-_105249308 | 18.91 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr13_+_91889626 | 18.73 |
ENSMUST00000022120.5
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr2_+_126398048 | 18.45 |
ENSMUST00000141482.3
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr9_-_103105638 | 18.22 |
ENSMUST00000126359.2
|
Trf
|
transferrin |
| chr18_-_38999755 | 17.62 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr19_-_8109346 | 17.61 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr8_+_105573693 | 17.41 |
ENSMUST00000055052.6
|
Ces2c
|
carboxylesterase 2C |
| chr3_-_79535966 | 17.31 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr8_+_13076024 | 17.26 |
ENSMUST00000033820.4
|
F7
|
coagulation factor VII |
| chr15_+_9335636 | 16.99 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr17_-_34247016 | 16.93 |
ENSMUST00000236627.2
ENSMUST00000237759.2 ENSMUST00000045467.14 ENSMUST00000114303.4 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr15_+_9279915 | 16.68 |
ENSMUST00000022861.9
|
Ugt3a1
|
UDP glycosyltransferases 3 family, polypeptide A1 |
| chr1_-_180021039 | 16.51 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr6_+_129510117 | 15.97 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr10_+_128089965 | 15.97 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr4_+_148686985 | 15.97 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr9_+_108539296 | 15.61 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr6_+_72575458 | 15.57 |
ENSMUST00000070597.13
ENSMUST00000176364.8 ENSMUST00000176168.3 |
Retsat
|
retinol saturase (all trans retinol 13,14 reductase) |
| chr10_-_127206300 | 15.34 |
ENSMUST00000026472.10
|
Inhbc
|
inhibin beta-C |
| chr7_-_99344832 | 15.30 |
ENSMUST00000145381.8
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr6_+_129510145 | 15.27 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr15_-_82278223 | 15.24 |
ENSMUST00000170255.2
|
Cyp2d11
|
cytochrome P450, family 2, subfamily d, polypeptide 11 |
| chr10_+_87694117 | 14.81 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr7_-_14172434 | 14.73 |
ENSMUST00000210396.2
ENSMUST00000168252.9 |
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr5_+_120614587 | 14.60 |
ENSMUST00000201684.4
ENSMUST00000066540.14 |
Sds
|
serine dehydratase |
| chr7_-_19432933 | 14.60 |
ENSMUST00000174355.8
ENSMUST00000172983.8 ENSMUST00000174710.2 ENSMUST00000003066.16 ENSMUST00000174064.9 |
Apoe
|
apolipoprotein E |
| chr15_+_3300249 | 14.50 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr11_-_69906171 | 14.46 |
ENSMUST00000018718.8
ENSMUST00000102574.10 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr19_-_44396092 | 14.32 |
ENSMUST00000041331.4
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr19_+_36532061 | 14.15 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr18_-_39000056 | 13.92 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr10_+_127612243 | 13.91 |
ENSMUST00000136223.2
ENSMUST00000052652.7 |
Rdh9
|
retinol dehydrogenase 9 |
| chr19_-_44017637 | 13.63 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr4_-_103072343 | 13.56 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr10_+_3490232 | 13.49 |
ENSMUST00000019896.5
|
Iyd
|
iodotyrosine deiodinase |
| chr10_+_127595639 | 13.41 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr9_-_106353792 | 13.37 |
ENSMUST00000214682.2
ENSMUST00000112479.9 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr12_-_104010690 | 13.15 |
ENSMUST00000043915.4
|
Serpina12
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr2_+_24970327 | 13.13 |
ENSMUST00000044078.10
ENSMUST00000114380.9 |
Entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr19_-_6899173 | 13.01 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr3_+_130411097 | 12.82 |
ENSMUST00000166187.8
ENSMUST00000072271.13 |
Etnppl
|
ethanolamine phosphate phospholyase |
| chr7_+_107166653 | 12.75 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr5_-_105387395 | 12.69 |
ENSMUST00000065588.7
|
Gbp10
|
guanylate-binding protein 10 |
| chr13_-_42001075 | 12.66 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr9_-_103165489 | 12.65 |
ENSMUST00000035163.10
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr9_-_103165423 | 12.48 |
ENSMUST00000123530.8
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr6_+_125297596 | 12.45 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr3_-_107925122 | 12.33 |
ENSMUST00000126593.3
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr7_+_107166925 | 12.25 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr8_+_105460627 | 12.16 |
ENSMUST00000034346.15
ENSMUST00000164182.3 |
Ces2a
|
carboxylesterase 2A |
| chr16_+_4825216 | 12.14 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr15_-_82291372 | 12.02 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr9_-_106353571 | 12.01 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr3_-_79536166 | 11.96 |
ENSMUST00000029386.14
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr19_-_6899121 | 11.83 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr7_+_140415170 | 11.61 |
ENSMUST00000211372.2
ENSMUST00000026554.11 ENSMUST00000185612.3 |
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr13_-_42001102 | 11.42 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr18_-_60860594 | 11.21 |
ENSMUST00000235795.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_+_121320008 | 11.12 |
ENSMUST00000166457.8
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr4_-_62069046 | 11.09 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr2_+_92205651 | 11.04 |
ENSMUST00000028650.9
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr2_-_91466739 | 11.04 |
ENSMUST00000111335.2
ENSMUST00000028681.15 |
F2
|
coagulation factor II |
| chr1_+_139429430 | 11.01 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr4_+_140970161 | 10.91 |
ENSMUST00000138096.8
ENSMUST00000006618.9 ENSMUST00000125392.8 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr10_+_128626772 | 10.87 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr11_-_78313043 | 10.83 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr5_-_38637474 | 10.73 |
ENSMUST00000143758.8
ENSMUST00000156272.8 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr17_-_84990360 | 10.59 |
ENSMUST00000066175.10
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr12_-_30423356 | 10.59 |
ENSMUST00000021004.14
|
Sntg2
|
syntrophin, gamma 2 |
| chr14_+_122771734 | 10.45 |
ENSMUST00000154206.8
ENSMUST00000038374.13 ENSMUST00000135578.8 |
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide |
| chr11_+_102652228 | 10.38 |
ENSMUST00000103081.10
ENSMUST00000068150.7 |
Adam11
|
a disintegrin and metallopeptidase domain 11 |
| chr17_-_57535003 | 10.36 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr18_+_74912268 | 10.35 |
ENSMUST00000041053.11
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
| chr11_+_73090270 | 10.34 |
ENSMUST00000006105.7
|
Shpk
|
sedoheptulokinase |
| chr17_+_84990541 | 10.22 |
ENSMUST00000045714.15
ENSMUST00000171915.2 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr1_+_88139678 | 10.19 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr3_-_67422821 | 10.17 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr1_-_180073492 | 10.11 |
ENSMUST00000010753.14
|
Psen2
|
presenilin 2 |
| chr2_-_32594043 | 10.09 |
ENSMUST00000143743.2
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr3_-_107925159 | 10.05 |
ENSMUST00000004140.11
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr7_+_140414837 | 10.04 |
ENSMUST00000106050.8
|
Urah
|
urate (5-hydroxyiso-) hydrolase |
| chr3_+_93462387 | 10.04 |
ENSMUST00000045756.14
|
S100a10
|
S100 calcium binding protein A10 (calpactin) |
| chr11_-_94932158 | 10.02 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr7_+_43093507 | 10.02 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr15_-_82264379 | 10.02 |
ENSMUST00000023083.9
|
Cyp2d22
|
cytochrome P450, family 2, subfamily d, polypeptide 22 |
| chrX_+_10118544 | 10.00 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr6_-_48549594 | 9.94 |
ENSMUST00000009425.7
ENSMUST00000204267.3 ENSMUST00000204930.3 ENSMUST00000204182.2 |
Rarres2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr6_-_119365632 | 9.93 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr5_-_37146266 | 9.91 |
ENSMUST00000166339.8
|
Wfs1
|
wolframin ER transmembrane glycoprotein |
| chr6_+_82029288 | 9.83 |
ENSMUST00000149023.2
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr16_+_4501934 | 9.62 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr13_-_41981812 | 9.60 |
ENSMUST00000223337.2
ENSMUST00000221691.2 |
Adtrp
|
androgen dependent TFPI regulating protein |
| chr5_+_31078775 | 9.60 |
ENSMUST00000201621.4
|
Khk
|
ketohexokinase |
| chr5_+_92719336 | 9.58 |
ENSMUST00000176621.8
ENSMUST00000175974.2 ENSMUST00000131166.9 ENSMUST00000176448.8 ENSMUST00000082382.8 |
Fam47e
|
family with sequence similarity 47, member E |
| chr13_-_41981893 | 9.56 |
ENSMUST00000137905.2
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr1_+_74324089 | 9.41 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr16_-_93726399 | 9.37 |
ENSMUST00000177648.8
ENSMUST00000142083.2 |
Cldn14
|
claudin 14 |
| chr2_-_91025441 | 9.35 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr3_+_130411294 | 9.34 |
ENSMUST00000163620.8
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr19_+_38995463 | 9.31 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chrX_+_10118600 | 9.27 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr1_+_36800874 | 9.27 |
ENSMUST00000027291.7
|
Zap70
|
zeta-chain (TCR) associated protein kinase |
| chr6_+_90310252 | 9.25 |
ENSMUST00000046128.12
ENSMUST00000164761.6 |
Uroc1
|
urocanase domain containing 1 |
| chr4_-_149569614 | 9.24 |
ENSMUST00000126896.2
ENSMUST00000105693.2 ENSMUST00000030845.13 |
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr15_+_101184488 | 9.23 |
ENSMUST00000229525.2
ENSMUST00000230525.2 |
Atg101
|
autophagy related 101 |
| chr5_-_38637624 | 9.21 |
ENSMUST00000067886.12
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr11_-_116089866 | 9.13 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr7_-_30643444 | 9.03 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr2_-_91025380 | 8.98 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr16_+_4825170 | 8.86 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr16_+_4825146 | 8.81 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr4_+_20007938 | 8.79 |
ENSMUST00000125799.8
ENSMUST00000121491.8 |
Ttpa
|
tocopherol (alpha) transfer protein |
| chr10_-_89342493 | 8.73 |
ENSMUST00000058126.15
ENSMUST00000105296.9 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr10_+_60113449 | 8.57 |
ENSMUST00000105465.8
ENSMUST00000179238.8 ENSMUST00000177779.8 ENSMUST00000004316.15 |
Psap
|
prosaposin |
| chr11_+_101258368 | 8.54 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr9_+_83807162 | 8.42 |
ENSMUST00000190637.7
ENSMUST00000034801.11 |
Bckdhb
|
branched chain ketoacid dehydrogenase E1, beta polypeptide |
| chr1_-_180073322 | 8.37 |
ENSMUST00000111104.2
|
Psen2
|
presenilin 2 |
| chr3_-_58433313 | 8.34 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr17_+_32904629 | 8.34 |
ENSMUST00000008801.7
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr2_-_154916367 | 8.34 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chr17_+_32904601 | 8.25 |
ENSMUST00000168171.8
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr12_-_84455764 | 8.15 |
ENSMUST00000120942.8
ENSMUST00000110272.9 |
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr3_-_121608859 | 8.12 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr12_+_108817043 | 8.11 |
ENSMUST00000057026.10
ENSMUST00000221080.2 |
Slc25a47
|
solute carrier family 25, member 47 |
| chr9_-_55419442 | 8.07 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr18_+_21077627 | 8.05 |
ENSMUST00000050004.3
|
Rnf125
|
ring finger protein 125 |
| chr11_+_4833186 | 8.01 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr7_-_4869160 | 7.99 |
ENSMUST00000064547.13
|
Isoc2b
|
isochorismatase domain containing 2b |
| chr11_+_115353290 | 7.94 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr7_-_45138188 | 7.93 |
ENSMUST00000011526.7
|
Dhdh
|
dihydrodiol dehydrogenase (dimeric) |
| chr15_-_83054369 | 7.93 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr1_+_163979384 | 7.92 |
ENSMUST00000086040.6
|
F5
|
coagulation factor V |
| chr9_-_121745354 | 7.91 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr9_+_107454114 | 7.88 |
ENSMUST00000112387.9
ENSMUST00000123005.8 ENSMUST00000010195.14 ENSMUST00000144392.2 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr15_-_100579813 | 7.87 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr19_-_47452557 | 7.78 |
ENSMUST00000111800.4
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr18_-_39051695 | 7.75 |
ENSMUST00000040647.11
|
Fgf1
|
fibroblast growth factor 1 |
| chr4_-_129142208 | 7.75 |
ENSMUST00000052602.6
|
C77080
|
expressed sequence C77080 |
| chr9_-_107546166 | 7.74 |
ENSMUST00000177567.8
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr2_-_91025492 | 7.72 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr10_+_76411474 | 7.69 |
ENSMUST00000001183.8
|
Ftcd
|
formiminotransferase cyclodeaminase |
| chr2_-_32321116 | 7.66 |
ENSMUST00000127961.3
ENSMUST00000136361.8 ENSMUST00000052119.14 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr2_-_91025208 | 7.64 |
ENSMUST00000111355.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr15_-_100579450 | 7.51 |
ENSMUST00000230740.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr16_-_17745999 | 7.47 |
ENSMUST00000003622.16
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr15_+_75088445 | 7.46 |
ENSMUST00000055719.8
|
Ly6g2
|
lymphocyte antigen 6 complex, locus G2 |
| chr4_+_155648157 | 7.38 |
ENSMUST00000105613.10
|
Nadk
|
NAD kinase |
| chr18_-_32271224 | 7.34 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pparg_Rxrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 38.3 | 153.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 23.9 | 71.7 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 14.0 | 41.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 13.1 | 65.6 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 12.3 | 49.1 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 11.5 | 46.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 10.0 | 29.9 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 9.3 | 111.0 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 8.4 | 33.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 7.6 | 22.8 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 7.5 | 22.4 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 6.6 | 66.3 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 6.3 | 44.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 6.2 | 191.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 6.2 | 18.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 5.9 | 52.8 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 5.8 | 51.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 5.4 | 168.3 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 5.2 | 10.3 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 5.1 | 15.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 5.1 | 30.4 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 4.9 | 14.6 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 4.8 | 33.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 4.8 | 14.3 | GO:1903699 | tarsal gland development(GO:1903699) |
| 4.4 | 22.0 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 4.3 | 184.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 4.2 | 8.3 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 4.1 | 98.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 3.6 | 29.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 3.5 | 52.2 | GO:0015747 | urate transport(GO:0015747) |
| 3.4 | 16.9 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 3.3 | 9.9 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 3.1 | 18.9 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 3.0 | 3.0 | GO:1904729 | regulation of intestinal lipid absorption(GO:1904729) |
| 3.0 | 23.7 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 2.9 | 8.8 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 2.9 | 17.3 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 2.8 | 8.5 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 2.7 | 16.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 2.7 | 18.9 | GO:0015886 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 2.7 | 16.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 2.7 | 8.0 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 2.6 | 7.9 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 2.6 | 7.9 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 2.6 | 10.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 2.5 | 37.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 2.5 | 10.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 2.4 | 17.1 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 2.4 | 9.6 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 2.3 | 9.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) fatty-acyl-CoA catabolic process(GO:0036115) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.3 | 9.0 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 2.2 | 11.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 2.2 | 6.7 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 2.2 | 11.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 2.2 | 8.8 | GO:0097017 | renal protein absorption(GO:0097017) |
| 2.2 | 8.7 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 2.1 | 22.9 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 2.0 | 8.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 2.0 | 6.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 2.0 | 33.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 2.0 | 31.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 1.9 | 7.7 | GO:0009804 | coumarin metabolic process(GO:0009804) coumarin catabolic process(GO:0046226) |
| 1.9 | 19.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 1.9 | 5.8 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 1.9 | 15.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.9 | 5.7 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.9 | 16.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 1.8 | 9.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 1.8 | 5.4 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.7 | 22.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.7 | 6.8 | GO:0090156 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 1.7 | 5.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 1.7 | 5.1 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.7 | 9.9 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 1.6 | 41.0 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 1.6 | 9.7 | GO:0043366 | beta selection(GO:0043366) |
| 1.6 | 9.6 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 1.6 | 6.4 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.6 | 6.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 1.6 | 10.9 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.6 | 20.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 1.5 | 4.6 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.5 | 7.6 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 1.5 | 4.5 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 1.5 | 36.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 1.5 | 7.5 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 1.5 | 13.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 1.5 | 16.3 | GO:0030242 | pexophagy(GO:0030242) |
| 1.4 | 4.3 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 1.4 | 4.3 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 1.4 | 4.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 1.4 | 18.2 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 1.4 | 5.6 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 1.4 | 5.5 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.4 | 21.6 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 1.4 | 5.4 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 1.3 | 4.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.3 | 10.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.3 | 27.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 1.3 | 5.3 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 1.3 | 6.6 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.3 | 3.8 | GO:1900239 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) regulation of phenotypic switching(GO:1900239) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 1.3 | 8.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 1.2 | 11.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.2 | 9.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.2 | 6.1 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 1.2 | 7.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.2 | 11.6 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.1 | 10.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.1 | 3.4 | GO:0006218 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 1.1 | 5.6 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 1.1 | 4.4 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 1.1 | 4.3 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 1.1 | 5.3 | GO:0046618 | drug export(GO:0046618) |
| 1.1 | 4.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 1.0 | 14.6 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 1.0 | 3.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 1.0 | 4.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 1.0 | 7.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.0 | 3.0 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 1.0 | 3.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 1.0 | 1.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.9 | 9.4 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.9 | 29.7 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.9 | 3.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.9 | 9.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.9 | 2.7 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.9 | 3.5 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.9 | 6.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.9 | 6.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.9 | 8.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.9 | 15.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.9 | 1.7 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.8 | 7.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.8 | 13.7 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.8 | 4.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.8 | 19.8 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.8 | 13.3 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.8 | 2.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.8 | 41.3 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.8 | 4.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.8 | 3.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.8 | 6.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.7 | 2.2 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.7 | 1.5 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.7 | 2.2 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.7 | 4.4 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.7 | 6.6 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.7 | 5.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.7 | 2.9 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.7 | 4.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.7 | 9.7 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.7 | 8.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.7 | 5.5 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.7 | 3.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.7 | 1.4 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.7 | 5.9 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.7 | 2.0 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) positive regulation of hindgut contraction(GO:0060450) |
| 0.6 | 1.9 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.6 | 5.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.6 | 4.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 7.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.6 | 2.9 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.6 | 2.9 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.6 | 2.9 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.6 | 4.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.6 | 6.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.6 | 1.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.6 | 15.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.6 | 1.7 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.5 | 1.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.5 | 4.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.5 | 3.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.5 | 1.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.5 | 1.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.5 | 3.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.5 | 1.5 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.5 | 10.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.5 | 4.6 | GO:1904152 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.5 | 0.5 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.5 | 5.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.5 | 3.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.5 | 5.0 | GO:1902739 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.5 | 2.5 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.5 | 9.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.5 | 1.5 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.5 | 5.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.5 | 2.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.5 | 1.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 2.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 2.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.5 | 6.0 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.5 | 8.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.5 | 1.4 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.4 | 2.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.4 | 1.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.4 | 10.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.4 | 2.6 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.4 | 5.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.4 | 1.7 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.4 | 8.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.4 | 1.6 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 4.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.4 | 5.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.4 | 0.4 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.4 | 2.0 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.4 | 13.6 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.4 | 2.4 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 0.8 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.4 | 1.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.4 | 1.5 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.4 | 3.9 | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.4 | 6.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.4 | 4.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.4 | 13.0 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.4 | 1.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.4 | 5.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.4 | 1.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.4 | 1.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.4 | 5.6 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 | 2.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 8.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.4 | 1.4 | GO:0051466 | paracrine signaling(GO:0038001) corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.4 | 2.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.3 | 1.4 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.3 | 11.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.3 | 8.1 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.3 | 2.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.3 | 1.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.3 | 0.7 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 | 7.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 5.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 2.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 5.7 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.3 | 0.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.3 | 3.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.3 | 0.6 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.3 | 5.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.3 | 0.9 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 5.0 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.3 | 12.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.3 | 4.6 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.3 | 1.5 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.3 | 5.3 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.3 | 4.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 4.8 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.3 | 2.8 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.3 | 23.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.3 | 4.5 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.3 | 1.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.3 | 4.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 1.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 1.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.3 | 0.8 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 0.5 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.3 | 1.1 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.3 | 2.1 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.3 | 12.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.3 | 1.8 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.3 | 0.5 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.3 | 14.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.3 | 7.7 | GO:0032094 | response to food(GO:0032094) |
| 0.2 | 2.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 3.9 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 1.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 1.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 2.8 | GO:0019682 | glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.2 | 5.1 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 0.9 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 5.0 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.2 | 4.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 0.9 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.2 | 2.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 3.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 4.9 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.2 | 0.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.2 | 1.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.2 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.2 | 3.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 1.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 4.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.9 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.2 | 1.5 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.2 | 3.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 1.6 | GO:1904684 | blood vessel maturation(GO:0001955) negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.2 | 1.0 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.2 | 4.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 0.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 0.4 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 | 1.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 0.8 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 7.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 5.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 0.8 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.2 | 2.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 2.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.2 | 10.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 0.7 | GO:0015800 | acidic amino acid transport(GO:0015800) |
| 0.2 | 1.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 1.5 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 1.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 2.7 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 5.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 2.9 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.2 | 1.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.2 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 0.5 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.2 | 6.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 0.5 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 3.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.2 | 1.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 9.1 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.2 | 4.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 6.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) response to nicotine(GO:0035094) |
| 0.2 | 1.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 0.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.6 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 0.3 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.2 | 1.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 1.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 0.9 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 2.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.9 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 4.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 2.9 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 1.0 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.1 | 4.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.7 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 2.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 2.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 5.2 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 1.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 7.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.3 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.3 | GO:0036145 | astrocyte chemotaxis(GO:0035700) dendritic cell homeostasis(GO:0036145) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.1 | 0.8 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.5 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.5 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 25.0 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.1 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 3.0 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 2.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 5.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 4.8 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 | 2.8 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.2 | GO:0003175 | tricuspid valve development(GO:0003175) |
| 0.1 | 0.3 | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000299) |
| 0.1 | 0.3 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 4.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 0.6 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.1 | 1.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 2.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 10.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 1.7 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 1.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.3 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.4 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.7 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 2.5 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.1 | 0.7 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.1 | 0.7 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 1.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 3.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 1.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.1 | 1.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.4 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.1 | 1.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 11.9 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.1 | 2.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.3 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.1 | 2.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 3.3 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 2.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 2.4 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 0.4 | GO:0060413 | atrial septum morphogenesis(GO:0060413) |
| 0.1 | 1.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 6.2 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 2.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 1.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 3.6 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 0.5 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.3 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.7 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.7 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 1.9 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 7.9 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 1.7 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 3.9 | GO:0097202 | activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.1 | 2.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 3.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 3.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 2.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.5 | GO:1903333 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.1 | 1.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.1 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 2.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.7 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.1 | 0.7 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 4.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 2.2 | GO:0035137 | hindlimb morphogenesis(GO:0035137) |
| 0.1 | 4.5 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.2 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.4 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 0.4 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.4 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 1.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 2.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 3.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.0 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 1.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 2.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 3.5 | GO:0071773 | response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 1.2 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.7 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 1.3 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.7 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 3.1 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.6 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.8 | GO:0048511 | rhythmic process(GO:0048511) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.3 | 157.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 11.8 | 47.4 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 4.2 | 16.7 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 3.4 | 33.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 3.3 | 10.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 3.2 | 38.1 | GO:0042627 | chylomicron(GO:0042627) |
| 2.6 | 23.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.5 | 12.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 2.4 | 7.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 2.3 | 6.9 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 2.2 | 24.2 | GO:0097433 | dense body(GO:0097433) |
| 2.0 | 59.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 1.6 | 24.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 1.6 | 4.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.5 | 7.7 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.5 | 7.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.5 | 4.4 | GO:0055087 | Ski complex(GO:0055087) |
| 1.4 | 6.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 1.3 | 22.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.1 | 32.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 1.1 | 4.3 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 1.1 | 5.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.1 | 5.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.1 | 5.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 1.0 | 6.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.0 | 10.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 1.0 | 14.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.9 | 15.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.9 | 2.7 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.9 | 173.9 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.9 | 5.1 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.8 | 5.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.8 | 21.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.8 | 15.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.8 | 3.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.8 | 40.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.7 | 9.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.7 | 151.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.7 | 60.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.7 | 17.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.6 | 1.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 3.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.6 | 30.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.6 | 6.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 3.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 1.7 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.5 | 2.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.5 | 4.6 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.5 | 5.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.4 | 6.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 25.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.4 | 1.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.4 | 12.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.4 | 6.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 1.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.4 | 5.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 2.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 1.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.4 | 4.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 10.9 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.3 | 1.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 3.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.3 | 3.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 9.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 2.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 10.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.3 | 1.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 5.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.3 | 6.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.3 | 0.9 | GO:0060473 | cortical granule(GO:0060473) |
| 0.3 | 6.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 1.5 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.3 | 3.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 1.5 | GO:0000802 | transverse filament(GO:0000802) |
| 0.3 | 5.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 3.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 9.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.3 | 3.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 1.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 5.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 24.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 5.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 6.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 48.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 4.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.2 | 1.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 1.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 8.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 94.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 369.1 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.2 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 1.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.6 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 7.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 1.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 6.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 1.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 1.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.2 | 1.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 1.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.5 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 3.3 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 3.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 21.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 6.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 10.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.4 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 17.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 2.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 2.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.9 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.1 | 0.3 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.1 | 1.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 4.4 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.1 | 2.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 5.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 1.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 81.3 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 4.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 4.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 3.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.3 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0035841 | growing cell tip(GO:0035838) new growing cell tip(GO:0035841) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 39.9 | 159.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 18.5 | 110.8 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 15.4 | 46.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 14.1 | 42.3 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 13.8 | 41.3 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 10.0 | 29.9 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 9.7 | 29.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 9.5 | 47.4 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 9.3 | 111.0 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 9.0 | 53.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 7.4 | 252.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 7.4 | 22.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 6.8 | 33.9 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 6.4 | 19.3 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 6.4 | 12.8 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 6.3 | 25.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 5.8 | 244.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 5.8 | 17.4 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 5.7 | 22.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 5.6 | 33.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 4.8 | 19.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 4.8 | 14.5 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 4.6 | 22.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 4.4 | 22.0 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 4.2 | 16.9 | GO:0070404 | NADH binding(GO:0070404) |
| 3.9 | 15.6 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 3.7 | 18.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 3.7 | 22.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 3.5 | 10.5 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 3.4 | 23.7 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 3.2 | 131.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 3.2 | 9.6 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 3.2 | 18.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 3.0 | 30.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 2.9 | 8.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 2.9 | 52.2 | GO:0008430 | selenium binding(GO:0008430) |
| 2.8 | 19.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 2.8 | 8.4 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 2.7 | 16.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 2.6 | 55.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 2.6 | 21.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 2.6 | 10.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 2.5 | 10.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 2.3 | 11.6 | GO:0035478 | chylomicron binding(GO:0035478) |
| 2.3 | 13.8 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 2.2 | 15.7 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 2.1 | 8.5 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 2.1 | 46.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 2.1 | 6.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 2.1 | 6.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 2.0 | 6.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 2.0 | 6.0 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 2.0 | 9.9 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 2.0 | 7.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.9 | 7.8 | GO:0018455 | alcohol dehydrogenase [NAD(P)+] activity(GO:0018455) |
| 1.9 | 7.7 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 1.9 | 7.7 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 1.9 | 7.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.9 | 11.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 1.8 | 18.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.8 | 7.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 1.7 | 8.7 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 1.7 | 5.2 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 1.7 | 11.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 1.7 | 29.7 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 1.6 | 21.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 1.6 | 8.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 1.6 | 9.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.6 | 4.8 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.6 | 31.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.5 | 7.5 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 1.5 | 7.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 1.4 | 12.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 1.4 | 39.1 | GO:0001848 | complement binding(GO:0001848) |
| 1.4 | 4.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 1.4 | 5.5 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.3 | 5.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 1.3 | 8.9 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.3 | 5.0 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.3 | 5.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.2 | 12.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 1.2 | 6.0 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 1.2 | 12.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.2 | 7.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.2 | 9.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.1 | 8.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.1 | 4.5 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 1.1 | 43.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.1 | 4.4 | GO:0004079 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 1.1 | 4.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.0 | 12.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.0 | 6.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.0 | 12.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 1.0 | 7.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.0 | 6.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 1.0 | 7.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.0 | 5.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 1.0 | 7.9 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 1.0 | 4.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 1.0 | 5.8 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.0 | 2.9 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.0 | 8.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.9 | 5.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.9 | 7.3 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) ether hydrolase activity(GO:0016803) |
| 0.9 | 11.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.9 | 2.7 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.9 | 1.8 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.9 | 5.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.9 | 10.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.9 | 18.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.9 | 2.6 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.9 | 3.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.8 | 34.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.8 | 14.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.8 | 7.4 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.8 | 3.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.8 | 13.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.8 | 3.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.8 | 7.7 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.7 | 2.2 | GO:0047651 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.7 | 2.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.7 | 4.4 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.7 | 13.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.7 | 2.9 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.7 | 2.9 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.7 | 4.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 11.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.7 | 2.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.7 | 4.6 | GO:0050610 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.7 | 9.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.6 | 3.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.6 | 5.6 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.6 | 3.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.6 | 7.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.6 | 6.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.6 | 1.8 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.6 | 6.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.6 | 3.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.6 | 8.8 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.5 | 1.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.5 | 2.7 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.5 | 5.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 41.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 5.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 4.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.5 | 6.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.5 | 1.5 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.5 | 2.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.5 | 29.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.5 | 1.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 1.5 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.5 | 10.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.5 | 5.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 6.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.5 | 4.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.5 | 4.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 5.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 0.9 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.5 | 23.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.5 | 1.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.4 | 3.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.4 | 8.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.4 | 1.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.4 | 2.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.4 | 10.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 5.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 1.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.4 | 36.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.4 | 9.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.4 | 2.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.4 | 3.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.4 | 14.4 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.4 | 6.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 5.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 1.1 | GO:0050354 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.4 | 1.8 | GO:0002046 | opsin binding(GO:0002046) |
| 0.4 | 2.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 5.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 38.7 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.4 | 40.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.4 | 1.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.3 | 1.0 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.3 | 0.7 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.3 | 2.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.3 | 4.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 0.9 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.3 | 0.9 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.3 | 3.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 16.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.3 | 2.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.3 | 4.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 4.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 1.4 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.3 | 2.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 1.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.3 | 0.8 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.3 | 6.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 33.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.3 | 1.1 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.3 | 20.8 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.3 | 2.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 10.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 4.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 8.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 5.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 4.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 5.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.5 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 2.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 13.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.7 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.2 | 5.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.2 | 8.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 5.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 1.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 2.8 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.2 | 1.6 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.2 | 18.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 10.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 0.6 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.2 | 2.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 3.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.7 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.2 | 1.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 7.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.9 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 1.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.2 | 9.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 0.3 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.2 | 1.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 8.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 28.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 12.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 8.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 2.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 4.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 14.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.8 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.1 | 0.9 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 2.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 2.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 8.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 2.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.6 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 0.4 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 2.0 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 2.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 3.9 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 0.2 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 5.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 4.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 4.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 1.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 1.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 1.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 5.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 2.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 7.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.7 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.3 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 2.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 15.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.0 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 2.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 1.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.8 | 44.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.6 | 50.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.6 | 16.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.6 | 42.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 10.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 31.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.4 | 5.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.3 | 4.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 5.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 73.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 9.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 3.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 1.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 1.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 5.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 13.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 5.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 9.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 5.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 9.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 6.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 8.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 3.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 5.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 8.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 2.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 13.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 125.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 8.0 | 24.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 7.4 | 148.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 5.5 | 66.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 5.3 | 84.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 4.2 | 46.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 3.5 | 114.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 3.2 | 41.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 2.5 | 35.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 2.4 | 41.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 2.2 | 44.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 2.2 | 23.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.8 | 23.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.8 | 26.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.7 | 15.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.7 | 19.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.6 | 22.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.4 | 30.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.4 | 24.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.4 | 40.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.3 | 22.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.3 | 30.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 1.2 | 12.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 1.2 | 21.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 1.1 | 12.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 1.1 | 21.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 1.0 | 15.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.9 | 7.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.9 | 9.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.9 | 11.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.8 | 61.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.8 | 13.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.7 | 6.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.7 | 56.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.7 | 23.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.7 | 71.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.7 | 7.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.7 | 15.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 87.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.5 | 12.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 4.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.5 | 6.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.4 | 6.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.4 | 7.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 5.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 11.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 7.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 5.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 12.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 12.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 5.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 11.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 10.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.3 | 3.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 5.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 14.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 5.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 7.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 10.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 5.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 5.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 7.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 3.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 4.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 6.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 3.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 4.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.2 | 5.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 5.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 4.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 2.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 6.1 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.2 | 7.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 5.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 1.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 5.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 5.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 13.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 7.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 3.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 4.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.5 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.1 | 2.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.5 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 5.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.9 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |