Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
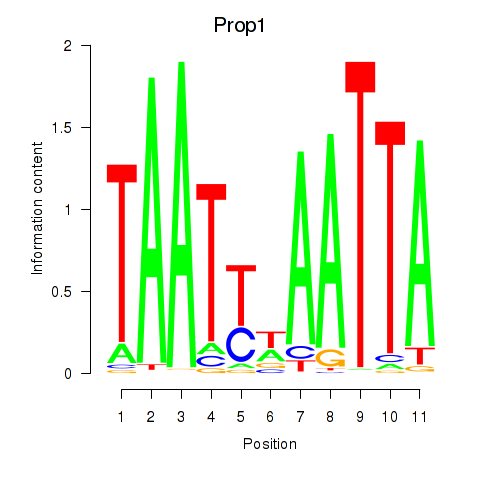
Results for Prop1
Z-value: 1.09
Transcription factors associated with Prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prop1
|
ENSMUSG00000044542.4 | paired like homeodomain factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prop1 | mm39_v1_chr11_-_50844572_50844596 | -0.42 | 1.0e-02 | Click! |
Activity profile of Prop1 motif
Sorted Z-values of Prop1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39275518 | 20.85 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr1_+_130754413 | 11.36 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr7_-_14180496 | 9.94 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr7_-_14180576 | 8.51 |
ENSMUST00000125941.3
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr3_+_138121245 | 7.49 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr1_-_130589349 | 6.31 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr1_-_130589321 | 6.12 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr10_-_115198093 | 4.94 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr5_+_9163244 | 4.71 |
ENSMUST00000198935.2
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr19_-_40175709 | 4.69 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr11_-_46597885 | 4.52 |
ENSMUST00000055102.13
ENSMUST00000125008.2 |
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr5_-_89605622 | 4.47 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr9_+_78137927 | 4.46 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr15_+_100202079 | 4.37 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr19_-_7943365 | 4.25 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr3_-_67422821 | 4.17 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr19_-_39637489 | 4.11 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr5_-_87682972 | 4.10 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr3_+_94284739 | 3.98 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr15_+_100202061 | 3.86 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr10_-_115197775 | 3.80 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr18_-_38999755 | 3.79 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr11_+_101932328 | 3.73 |
ENSMUST00000123895.8
ENSMUST00000017453.12 ENSMUST00000107163.9 ENSMUST00000107164.3 |
Cd300lg
|
CD300 molecule like family member G |
| chr3_+_94284812 | 3.70 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr4_+_134124691 | 3.61 |
ENSMUST00000105870.8
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr15_+_100202021 | 3.59 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr17_+_85335775 | 3.44 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr1_+_167426019 | 3.40 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr9_+_78164402 | 3.32 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr3_+_122688721 | 2.93 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr6_+_149043011 | 2.90 |
ENSMUST00000179873.8
ENSMUST00000047531.16 ENSMUST00000111548.8 ENSMUST00000111547.2 ENSMUST00000134306.8 ENSMUST00000147934.4 |
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr7_+_132212349 | 2.82 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr12_+_9079966 | 2.64 |
ENSMUST00000085741.2
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr3_+_137923521 | 2.59 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr19_+_11514132 | 2.59 |
ENSMUST00000025581.7
|
Ms4a4d
|
membrane-spanning 4-domains, subfamily A, member 4D |
| chr4_+_115458172 | 2.48 |
ENSMUST00000084342.6
|
Cyp4a32
|
cytochrome P450, family 4, subfamily a, polypeptide 32 |
| chr7_+_100970287 | 2.42 |
ENSMUST00000032927.14
|
Stard10
|
START domain containing 10 |
| chr17_+_64907697 | 2.38 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr1_+_167425953 | 2.32 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr11_-_43792013 | 2.23 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr13_+_24511387 | 2.08 |
ENSMUST00000224953.2
ENSMUST00000050859.13 ENSMUST00000167746.8 ENSMUST00000224819.2 |
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr14_-_45715308 | 2.04 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr6_+_149043136 | 2.02 |
ENSMUST00000166416.8
ENSMUST00000111551.2 |
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr12_+_9080014 | 2.01 |
ENSMUST00000219488.2
ENSMUST00000219470.2 |
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr3_+_122213420 | 1.99 |
ENSMUST00000029766.9
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr13_-_56696310 | 1.99 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr18_-_39000056 | 1.95 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr9_+_53212871 | 1.84 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr4_+_98919183 | 1.82 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr6_-_83098255 | 1.80 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chr2_-_110136074 | 1.80 |
ENSMUST00000046233.9
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr5_+_130398261 | 1.74 |
ENSMUST00000086029.10
|
Caln1
|
calneuron 1 |
| chr17_-_45910529 | 1.72 |
ENSMUST00000171847.8
ENSMUST00000166633.8 ENSMUST00000169729.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr12_-_79237722 | 1.71 |
ENSMUST00000085254.7
|
Rdh11
|
retinol dehydrogenase 11 |
| chr11_-_12362136 | 1.67 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr9_+_66853343 | 1.66 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr2_-_134396268 | 1.64 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr1_-_106687457 | 1.61 |
ENSMUST00000010049.6
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr6_-_115569504 | 1.61 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr7_+_101467512 | 1.60 |
ENSMUST00000008090.11
|
Phox2a
|
paired-like homeobox 2a |
| chr9_+_65538352 | 1.59 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr12_-_57592907 | 1.57 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr5_-_90371816 | 1.55 |
ENSMUST00000118816.6
ENSMUST00000048363.10 |
Cox18
|
cytochrome c oxidase assembly protein 18 |
| chrX_-_99638466 | 1.55 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr18_-_3299536 | 1.54 |
ENSMUST00000129435.8
ENSMUST00000122958.8 |
Crem
|
cAMP responsive element modulator |
| chr12_+_85157607 | 1.54 |
ENSMUST00000053811.10
|
Dlst
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr18_+_32200781 | 1.53 |
ENSMUST00000025243.5
ENSMUST00000212675.2 |
Iws1
|
IWS1, SUPT6 interacting protein |
| chr13_-_4659120 | 1.51 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr1_-_24044688 | 1.48 |
ENSMUST00000027338.4
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr7_-_30555592 | 1.45 |
ENSMUST00000185748.2
ENSMUST00000094583.2 |
Ffar3
|
free fatty acid receptor 3 |
| chr9_+_80072361 | 1.45 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr11_-_59054107 | 1.44 |
ENSMUST00000069631.3
|
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr8_+_13110921 | 1.44 |
ENSMUST00000211363.2
ENSMUST00000033822.4 |
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr4_+_95445731 | 1.41 |
ENSMUST00000079223.11
ENSMUST00000177394.8 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chrM_+_10167 | 1.38 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr4_+_100336003 | 1.33 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr4_-_14826587 | 1.32 |
ENSMUST00000117268.9
ENSMUST00000236953.2 |
Otud6b
|
OTU domain containing 6B |
| chr19_-_39801188 | 1.30 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr4_-_108075119 | 1.26 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr4_+_109835224 | 1.25 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr5_+_42225303 | 1.24 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr4_-_82423511 | 1.23 |
ENSMUST00000050872.15
ENSMUST00000064770.9 |
Nfib
|
nuclear factor I/B |
| chr11_-_49004584 | 1.22 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr12_+_59176543 | 1.21 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr4_-_82423985 | 1.18 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr5_-_31265562 | 1.17 |
ENSMUST00000201396.2
ENSMUST00000202740.4 |
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chrX_+_109857866 | 1.17 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chrX_+_37689503 | 1.16 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr1_+_134217727 | 1.15 |
ENSMUST00000027730.6
|
Myog
|
myogenin |
| chr2_+_81883566 | 1.14 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr9_-_59260713 | 1.14 |
ENSMUST00000026265.8
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr3_-_92031247 | 1.13 |
ENSMUST00000070284.4
|
Prr9
|
proline rich 9 |
| chr14_+_73411249 | 1.11 |
ENSMUST00000166875.2
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr3_+_145827410 | 1.09 |
ENSMUST00000039450.5
|
Mcoln3
|
mucolipin 3 |
| chr7_+_6733561 | 1.08 |
ENSMUST00000200535.6
|
Usp29
|
ubiquitin specific peptidase 29 |
| chr10_+_79977291 | 1.05 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr17_-_25394445 | 1.05 |
ENSMUST00000224277.2
|
Percc1
|
proline and glutamate rich with coiled coil 1 |
| chr4_-_12087911 | 1.05 |
ENSMUST00000050686.10
|
Tmem67
|
transmembrane protein 67 |
| chr12_+_59176506 | 1.03 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr8_+_48275178 | 1.03 |
ENSMUST00000079639.3
|
Cldn24
|
claudin 24 |
| chr18_+_38429792 | 1.02 |
ENSMUST00000237211.2
|
Rnf14
|
ring finger protein 14 |
| chrX_+_37861548 | 1.02 |
ENSMUST00000050744.6
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr5_+_92285748 | 1.01 |
ENSMUST00000031355.10
ENSMUST00000202155.2 |
Uso1
|
USO1 vesicle docking factor |
| chr2_+_124978518 | 1.01 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr7_+_6733684 | 1.01 |
ENSMUST00000197117.5
|
Usp29
|
ubiquitin specific peptidase 29 |
| chrM_+_9870 | 1.01 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr3_-_116506294 | 1.00 |
ENSMUST00000029569.9
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr3_-_141687987 | 0.99 |
ENSMUST00000029948.15
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr1_-_57008986 | 0.98 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr1_+_37911415 | 0.98 |
ENSMUST00000142670.2
|
Lipt1
|
lipoyltransferase 1 |
| chr11_+_43046476 | 0.97 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr4_-_82423944 | 0.97 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr16_+_56024676 | 0.97 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr14_-_33996185 | 0.96 |
ENSMUST00000227006.2
|
Shld2
|
shieldin complex subunit 2 |
| chr16_-_64591509 | 0.96 |
ENSMUST00000076991.7
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene |
| chr1_+_37911272 | 0.96 |
ENSMUST00000041621.5
|
Lipt1
|
lipoyltransferase 1 |
| chr9_-_8134295 | 0.95 |
ENSMUST00000037397.8
|
Cep126
|
centrosomal protein 126 |
| chr15_-_96947963 | 0.94 |
ENSMUST00000230907.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr4_-_41517326 | 0.94 |
ENSMUST00000030152.13
ENSMUST00000095126.5 |
1110017D15Rik
|
RIKEN cDNA 1110017D15 gene |
| chr16_-_19341016 | 0.93 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr6_-_136150491 | 0.93 |
ENSMUST00000111905.8
ENSMUST00000152012.8 ENSMUST00000143943.8 ENSMUST00000125905.2 |
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr6_+_86342622 | 0.91 |
ENSMUST00000071492.9
|
Fam136a
|
family with sequence similarity 136, member A |
| chr6_+_3993774 | 0.90 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr3_-_10396418 | 0.90 |
ENSMUST00000191670.6
ENSMUST00000065938.15 ENSMUST00000118410.8 |
Impa1
|
inositol (myo)-1(or 4)-monophosphatase 1 |
| chr8_+_22682816 | 0.89 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr2_+_86655007 | 0.89 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr3_-_72965136 | 0.88 |
ENSMUST00000059407.9
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr6_+_40619913 | 0.88 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr16_-_44153288 | 0.87 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr18_+_38429858 | 0.87 |
ENSMUST00000171461.3
ENSMUST00000235811.2 |
Rnf14
|
ring finger protein 14 |
| chr11_+_100978103 | 0.86 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chr10_+_69761630 | 0.86 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr16_+_62635039 | 0.85 |
ENSMUST00000055557.6
|
Stx19
|
syntaxin 19 |
| chr11_-_54140462 | 0.83 |
ENSMUST00000019060.6
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chrX_+_102674181 | 0.82 |
ENSMUST00000033692.9
|
Zcchc13
|
zinc finger, CCHC domain containing 13 |
| chr6_+_115398996 | 0.82 |
ENSMUST00000000450.5
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chrX_-_133177638 | 0.81 |
ENSMUST00000113252.8
|
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr17_-_15163362 | 0.81 |
ENSMUST00000238668.2
ENSMUST00000228330.2 |
Wdr27
|
WD repeat domain 27 |
| chr5_-_138617952 | 0.80 |
ENSMUST00000063262.11
ENSMUST00000085852.11 ENSMUST00000110905.9 |
Zfp68
|
zinc finger protein 68 |
| chr1_+_172777976 | 0.77 |
ENSMUST00000215254.2
|
Olfr16
|
olfactory receptor 16 |
| chr7_+_6442835 | 0.76 |
ENSMUST00000168341.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr12_-_104439589 | 0.76 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr5_+_104318542 | 0.75 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr17_+_29487881 | 0.75 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr9_+_100479732 | 0.75 |
ENSMUST00000124487.8
|
Stag1
|
stromal antigen 1 |
| chr6_+_41512010 | 0.74 |
ENSMUST00000103288.2
|
Trbj1-5
|
T cell receptor beta joining 1-5 |
| chrX_+_139808351 | 0.73 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr2_+_121786892 | 0.71 |
ENSMUST00000110578.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr11_-_99313078 | 0.71 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr13_+_67981349 | 0.71 |
ENSMUST00000222626.2
ENSMUST00000060609.8 |
Gm10037
|
predicted gene 10037 |
| chr18_+_38430015 | 0.71 |
ENSMUST00000236319.2
|
Rnf14
|
ring finger protein 14 |
| chrX_-_133177717 | 0.70 |
ENSMUST00000087541.12
ENSMUST00000087540.4 |
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr7_-_12829100 | 0.70 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr7_-_24423715 | 0.70 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chrM_+_11735 | 0.70 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr2_+_124978612 | 0.69 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr2_-_89671899 | 0.69 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr2_+_121787131 | 0.68 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr2_+_27055245 | 0.68 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr3_+_84500854 | 0.68 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr13_+_36301830 | 0.67 |
ENSMUST00000099582.4
ENSMUST00000224611.2 |
Fars2
|
phenylalanine-tRNA synthetase 2 (mitochondrial) |
| chr2_-_71377088 | 0.67 |
ENSMUST00000024159.8
|
Dlx2
|
distal-less homeobox 2 |
| chr2_+_88505972 | 0.67 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr14_+_75693396 | 0.66 |
ENSMUST00000164848.3
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr7_+_140521450 | 0.66 |
ENSMUST00000164580.3
ENSMUST00000079403.11 |
Pgghg
|
protein glucosylgalactosylhydroxylysine glucosidase |
| chr16_-_44153498 | 0.65 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr4_-_3938352 | 0.65 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr16_-_16962256 | 0.64 |
ENSMUST00000115711.10
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr18_-_78640066 | 0.64 |
ENSMUST00000235389.2
ENSMUST00000237674.2 |
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chrM_+_7779 | 0.63 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr3_-_145813802 | 0.63 |
ENSMUST00000160285.2
|
Dnai3
|
dynein axonemal intermediate chain 3 |
| chr13_-_21798192 | 0.63 |
ENSMUST00000051874.6
|
Olfr1362
|
olfactory receptor 1362 |
| chr18_+_3382968 | 0.62 |
ENSMUST00000025073.12
|
Cul2
|
cullin 2 |
| chr15_+_98468885 | 0.62 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr7_+_106740521 | 0.62 |
ENSMUST00000210474.2
|
Olfr716
|
olfactory receptor 716 |
| chr9_+_118307412 | 0.62 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr11_-_117764258 | 0.61 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr1_-_163552693 | 0.61 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr12_-_113928438 | 0.61 |
ENSMUST00000103478.4
|
Ighv3-1
|
immunoglobulin heavy variable 3-1 |
| chr3_-_116506345 | 0.60 |
ENSMUST00000169530.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr7_-_24145107 | 0.60 |
ENSMUST00000205776.2
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr11_+_64869850 | 0.60 |
ENSMUST00000071891.12
ENSMUST00000108697.8 ENSMUST00000101049.9 |
Elac2
|
elaC ribonuclease Z 2 |
| chr12_-_84664001 | 0.60 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr2_+_74557418 | 0.59 |
ENSMUST00000111980.4
|
Hoxd4
|
homeobox D4 |
| chr13_+_19427015 | 0.59 |
ENSMUST00000198663.5
ENSMUST00000103559.3 |
Trgv3
|
T cell receptor gamma, variable 3 |
| chr15_+_98350469 | 0.59 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chrM_+_2743 | 0.58 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr18_-_9282754 | 0.57 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4 |
| chr7_-_45343785 | 0.57 |
ENSMUST00000040636.9
|
Sec1
|
secretory blood group 1 |
| chr18_-_66993567 | 0.56 |
ENSMUST00000057942.4
|
Mc4r
|
melanocortin 4 receptor |
| chr9_+_118307250 | 0.55 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr13_-_43634695 | 0.54 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr6_+_37847721 | 0.54 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr14_-_51433380 | 0.53 |
ENSMUST00000051274.2
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr16_-_92155762 | 0.53 |
ENSMUST00000166707.3
|
Kcne1
|
potassium voltage-gated channel, Isk-related subfamily, member 1 |
| chr1_+_87731360 | 0.52 |
ENSMUST00000177757.2
ENSMUST00000077772.12 |
Sag
|
S-antigen, retina and pineal gland (arrestin) |
| chr16_-_94023976 | 0.51 |
ENSMUST00000227698.2
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
| chr11_+_84020475 | 0.51 |
ENSMUST00000133811.3
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr3_+_41697046 | 0.51 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr4_-_119349760 | 0.51 |
ENSMUST00000049994.8
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prop1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 2.5 | 10.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.6 | 4.9 | GO:1904735 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 1.4 | 4.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.9 | 30.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.8 | 3.4 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 5.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.7 | 2.2 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.7 | 7.7 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.5 | 1.6 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.5 | 1.6 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.5 | 1.6 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.5 | 2.1 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 | 1.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 1.7 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 1.1 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.4 | 1.4 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.3 | 1.6 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.3 | 1.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 1.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.3 | 1.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.3 | 4.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 2.4 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.3 | 1.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 0.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 1.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 4.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.7 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.2 | 0.7 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 0.9 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 2.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.8 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.6 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.2 | 1.0 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 1.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 1.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 1.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) regulation of mRNA export from nucleus(GO:0010793) |
| 0.2 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.5 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.2 | 2.0 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 4.5 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 1.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.7 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 5.7 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.5 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.1 | 0.4 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.1 | 0.6 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 1.6 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.5 | GO:2000292 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 1.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 4.5 | GO:0097286 | iron ion import(GO:0097286) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.2 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 1.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.8 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.5 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.6 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 0.8 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 1.6 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.7 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.9 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.6 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.4 | GO:0034465 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.1 | 9.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 2.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 1.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 2.2 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.6 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 1.5 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 1.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.4 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.3 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.8 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.4 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.6 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.4 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.6 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 1.3 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 2.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 1.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.3 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 3.2 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.7 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.3 | GO:0071158 | ectoderm development(GO:0007398) positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 0.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.3 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 1.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 2.1 | GO:0006865 | amino acid transport(GO:0006865) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 2.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 11.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 1.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 0.7 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.2 | 1.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 1.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 0.7 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 3.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 2.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.4 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 2.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.9 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 2.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 2.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 8.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 2.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 3.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 3.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 6.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 3.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 20.8 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 3.4 | 10.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.3 | 7.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.1 | 4.5 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.0 | 4.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.9 | 11.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.9 | 4.5 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.8 | 2.4 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.7 | 2.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.7 | 2.1 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.6 | 1.9 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.6 | 5.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.6 | 2.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 1.5 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.5 | 1.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.4 | 1.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.4 | 1.5 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.4 | 11.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 2.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 3.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 5.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 3.6 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.3 | 1.5 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 1.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 4.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 0.7 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 1.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 0.6 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.2 | 0.6 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 0.9 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 4.5 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 0.6 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 1.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.9 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.5 | GO:0004078 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 3.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.5 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.6 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 3.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.6 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 1.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.3 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 2.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 1.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 2.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 2.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 3.4 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 4.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 2.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 3.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 2.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 10.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 4.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 5.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 14.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 6.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 3.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 4.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |