Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
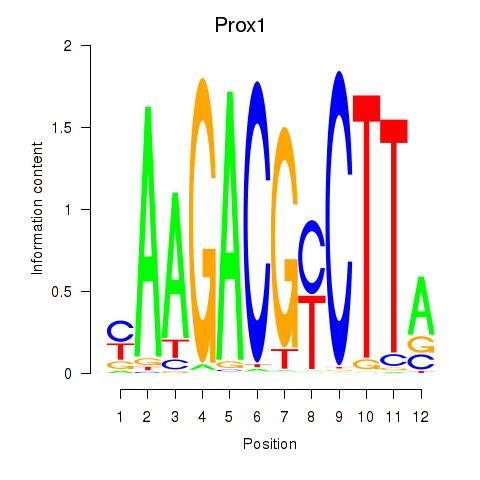
Results for Prox1
Z-value: 1.07
Transcription factors associated with Prox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prox1
|
ENSMUSG00000010175.14 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prox1 | mm39_v1_chr1_-_189901596_189901723 | 0.04 | 8.0e-01 | Click! |
Activity profile of Prox1 motif
Sorted Z-values of Prox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_137157824 | 18.08 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr14_-_51384236 | 17.26 |
ENSMUST00000080126.4
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr19_-_15901919 | 9.31 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr7_+_141995545 | 4.71 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr4_+_130640611 | 4.56 |
ENSMUST00000156225.8
ENSMUST00000156742.8 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr6_-_131365380 | 4.09 |
ENSMUST00000032309.13
ENSMUST00000087865.4 |
Ybx3
|
Y box protein 3 |
| chr4_-_133694607 | 4.03 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_-_137137088 | 3.66 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr4_-_133694543 | 3.58 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_+_130640436 | 3.54 |
ENSMUST00000151698.8
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr4_+_140428777 | 3.07 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr17_-_35827676 | 3.05 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr11_+_4936824 | 2.93 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr19_+_46044972 | 2.90 |
ENSMUST00000111899.8
ENSMUST00000099392.10 ENSMUST00000062322.11 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr2_+_36120438 | 2.90 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr15_-_86070338 | 2.84 |
ENSMUST00000044332.16
|
Cerk
|
ceramide kinase |
| chr17_+_29538157 | 2.61 |
ENSMUST00000114699.9
ENSMUST00000155348.3 ENSMUST00000234618.2 |
Pi16
|
peptidase inhibitor 16 |
| chr12_+_111409087 | 2.33 |
ENSMUST00000109792.8
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr5_+_139238089 | 2.13 |
ENSMUST00000130326.8
|
Get4
|
golgi to ER traffic protein 4 |
| chr2_-_26800581 | 1.99 |
ENSMUST00000015920.12
ENSMUST00000139815.2 ENSMUST00000102899.10 |
Med22
|
mediator complex subunit 22 |
| chr3_+_153549846 | 1.75 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr14_-_56181993 | 1.74 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr5_+_139238069 | 1.74 |
ENSMUST00000026976.12
|
Get4
|
golgi to ER traffic protein 4 |
| chr4_+_132903646 | 1.72 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chrX_-_73009933 | 1.70 |
ENSMUST00000114372.3
ENSMUST00000033761.13 |
Hcfc1
|
host cell factor C1 |
| chr17_+_28059129 | 1.69 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr12_-_83534482 | 1.67 |
ENSMUST00000177959.8
ENSMUST00000178756.8 |
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr19_-_8691797 | 1.62 |
ENSMUST00000206797.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr7_-_126460820 | 1.58 |
ENSMUST00000129812.2
ENSMUST00000106342.8 |
Ino80e
|
INO80 complex subunit E |
| chr8_+_89015705 | 1.54 |
ENSMUST00000171456.9
|
Adcy7
|
adenylate cyclase 7 |
| chr12_-_40273173 | 1.44 |
ENSMUST00000001672.12
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr17_+_28059099 | 1.42 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr8_-_70929555 | 1.38 |
ENSMUST00000066597.13
ENSMUST00000210250.2 ENSMUST00000209415.2 ENSMUST00000166976.3 |
Klhl26
|
kelch-like 26 |
| chr17_+_28059036 | 1.30 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr19_-_8691220 | 1.27 |
ENSMUST00000010239.6
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr18_+_10617773 | 1.24 |
ENSMUST00000002551.5
ENSMUST00000234207.2 |
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr18_-_68562385 | 1.15 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr4_+_117692583 | 1.15 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr5_-_124492734 | 1.15 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr6_+_58617521 | 1.04 |
ENSMUST00000145161.8
ENSMUST00000203146.3 ENSMUST00000114294.8 ENSMUST00000204948.2 |
Abcg2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr8_+_31601837 | 1.01 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr5_+_139197689 | 0.94 |
ENSMUST00000148772.8
ENSMUST00000110882.8 |
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr2_+_26800757 | 0.91 |
ENSMUST00000102898.5
|
Rpl7a
|
ribosomal protein L7A |
| chr11_-_8614497 | 0.90 |
ENSMUST00000020695.13
|
Tns3
|
tensin 3 |
| chr3_+_89738654 | 0.89 |
ENSMUST00000050401.6
|
She
|
src homology 2 domain-containing transforming protein E |
| chr10_-_80649315 | 0.89 |
ENSMUST00000181039.8
ENSMUST00000180438.2 |
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr5_+_24305577 | 0.87 |
ENSMUST00000030841.10
ENSMUST00000163409.5 |
Klhl7
|
kelch-like 7 |
| chr6_+_29361408 | 0.77 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr7_+_127376267 | 0.76 |
ENSMUST00000144406.8
|
Setd1a
|
SET domain containing 1A |
| chr6_+_60921456 | 0.75 |
ENSMUST00000129603.4
ENSMUST00000204333.2 |
Mmrn1
|
multimerin 1 |
| chr6_-_42350188 | 0.71 |
ENSMUST00000073387.5
ENSMUST00000204357.2 |
Epha1
|
Eph receptor A1 |
| chr18_-_67378886 | 0.66 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr11_-_115824290 | 0.54 |
ENSMUST00000021097.10
|
Recql5
|
RecQ protein-like 5 |
| chr2_+_109848224 | 0.49 |
ENSMUST00000150183.9
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr18_+_37864045 | 0.44 |
ENSMUST00000192535.2
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chrX_-_42363663 | 0.41 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr17_-_83821716 | 0.36 |
ENSMUST00000025095.9
ENSMUST00000167741.9 |
Cox7a2l
|
cytochrome c oxidase subunit 7A2 like |
| chr2_-_92290128 | 0.30 |
ENSMUST00000067631.7
|
Slc35c1
|
solute carrier family 35, member C1 |
| chr2_-_92290054 | 0.29 |
ENSMUST00000136718.2
|
Slc35c1
|
solute carrier family 35, member C1 |
| chr2_+_26899935 | 0.27 |
ENSMUST00000114005.9
ENSMUST00000114004.8 ENSMUST00000114006.8 ENSMUST00000114007.8 ENSMUST00000133807.2 |
Cacfd1
|
calcium channel flower domain containing 1 |
| chr9_-_64633865 | 0.23 |
ENSMUST00000168366.2
|
Rab11a
|
RAB11A, member RAS oncogene family |
| chr7_+_28416270 | 0.22 |
ENSMUST00000108279.9
|
Fbxo17
|
F-box protein 17 |
| chr2_+_86128161 | 0.20 |
ENSMUST00000054746.5
|
Olfr1052
|
olfactory receptor 1052 |
| chr5_+_62968993 | 0.18 |
ENSMUST00000238474.2
|
Dthd1
|
death domain containing 1 |
| chr7_+_23781311 | 0.17 |
ENSMUST00000207002.2
ENSMUST00000068975.6 ENSMUST00000203854.3 |
Zfp180
|
zinc finger protein 180 |
| chr1_+_78794475 | 0.13 |
ENSMUST00000057262.8
ENSMUST00000187432.2 |
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr9_+_109760931 | 0.10 |
ENSMUST00000165876.8
|
Map4
|
microtubule-associated protein 4 |
| chr11_+_115671523 | 0.03 |
ENSMUST00000239299.2
|
Tmem94
|
transmembrane protein 94 |
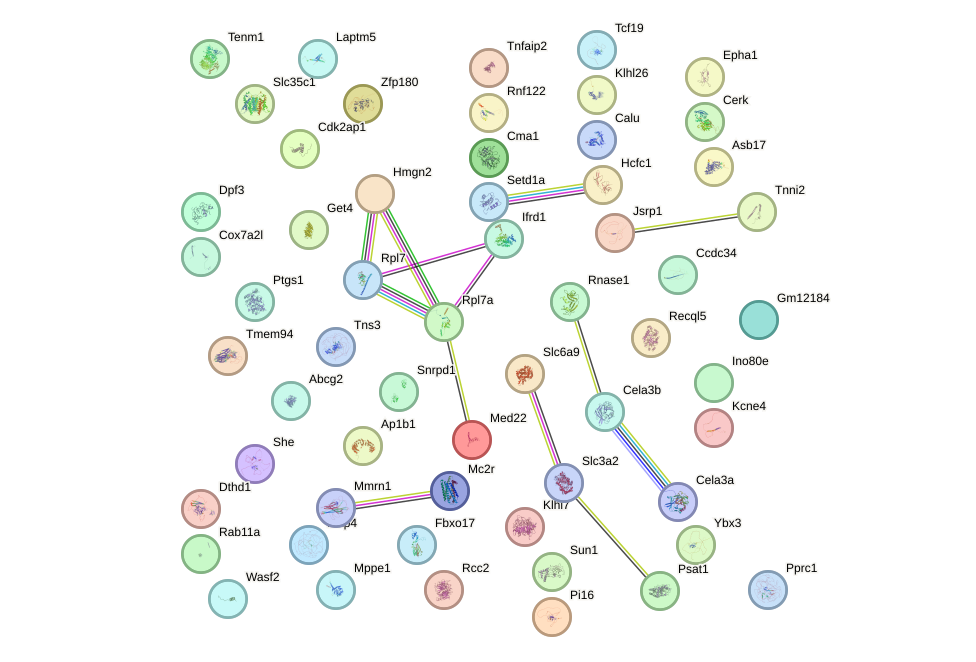
Network of associatons between targets according to the STRING database.
First level regulatory network of Prox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.0 | 4.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 1.0 | 3.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.6 | 2.9 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.6 | 2.9 | GO:0060356 | leucine import(GO:0060356) |
| 0.5 | 4.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.5 | 3.9 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.4 | 1.7 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.4 | 1.7 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.4 | 1.2 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.3 | 1.0 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 7.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 17.3 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 2.6 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 2.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.9 | GO:0090292 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.5 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 21.6 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 1.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 5.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 3.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 2.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.4 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 1.7 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 7.7 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 2.8 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 1.2 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.9 | GO:0048286 | lung alveolus development(GO:0048286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 4.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 4.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 2.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 4.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 8.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 17.3 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 1.1 | 4.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.9 | 2.8 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.5 | 4.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 1.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.2 | 0.8 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 5.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.7 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 23.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 2.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 2.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 3.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 2.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 20.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 2.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 5.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 3.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |