Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Prrx1_Isx_Prrxl1
Z-value: 0.47
Transcription factors associated with Prrx1_Isx_Prrxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
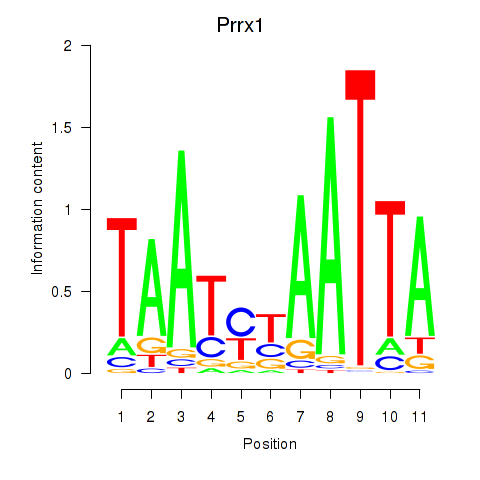
Prrx1
|
ENSMUSG00000026586.17 | paired related homeobox 1 |
|
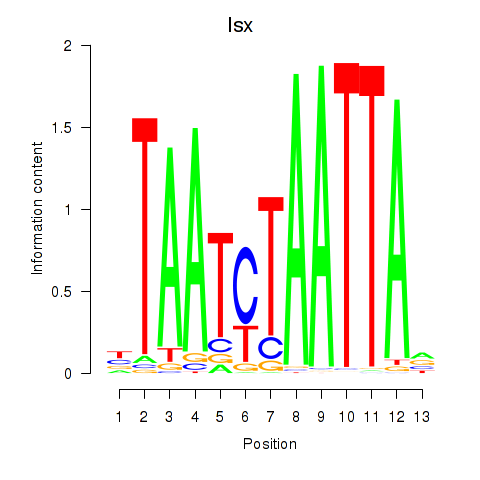
Isx
|
ENSMUSG00000031621.10 | intestine specific homeobox |
|
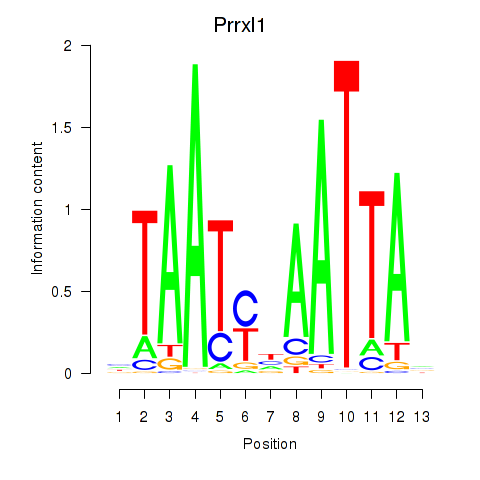
Prrxl1
|
ENSMUSG00000041730.15 | paired related homeobox protein-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isx | mm39_v1_chr8_+_75599801_75599855 | 0.28 | 1.0e-01 | Click! |
| Prrx1 | mm39_v1_chr1_-_163141230_163141246 | -0.23 | 1.7e-01 | Click! |
| Prrxl1 | mm39_v1_chr14_+_32321341_32321498 | -0.18 | 2.9e-01 | Click! |
Activity profile of Prrx1_Isx_Prrxl1 motif
Sorted Z-values of Prrx1_Isx_Prrxl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_130754413 | 1.51 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr13_+_4283729 | 1.24 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr7_-_14180496 | 1.24 |
ENSMUST00000063509.11
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chrX_-_93256244 | 1.12 |
ENSMUST00000113922.2
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr7_-_14180576 | 1.11 |
ENSMUST00000125941.3
|
Sult2a8
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
| chr12_+_9079966 | 0.97 |
ENSMUST00000085741.2
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr3_+_59989282 | 0.81 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr10_+_82695013 | 0.76 |
ENSMUST00000020484.9
ENSMUST00000219962.3 |
Txnrd1
|
thioredoxin reductase 1 |
| chr1_-_130589349 | 0.75 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr1_-_130589321 | 0.75 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr15_+_100202079 | 0.74 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr3_+_138121245 | 0.72 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr15_+_65682066 | 0.71 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr11_-_12362136 | 0.69 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr10_+_82695055 | 0.68 |
ENSMUST00000218694.2
|
Txnrd1
|
thioredoxin reductase 1 |
| chr15_+_100202061 | 0.67 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr19_-_8109346 | 0.65 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr15_+_100202021 | 0.65 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr9_+_78137927 | 0.64 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr12_+_9080014 | 0.61 |
ENSMUST00000219488.2
ENSMUST00000219470.2 |
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr1_-_150341911 | 0.60 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr18_-_43610829 | 0.59 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr4_+_103000248 | 0.57 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chr18_-_38999755 | 0.56 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr18_+_3382968 | 0.55 |
ENSMUST00000025073.12
|
Cul2
|
cullin 2 |
| chrM_+_11735 | 0.55 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_-_87682972 | 0.52 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr5_+_9163244 | 0.52 |
ENSMUST00000198935.2
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr8_+_22682816 | 0.51 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr1_-_162726234 | 0.47 |
ENSMUST00000111510.8
ENSMUST00000045902.13 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr6_+_37847721 | 0.46 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr7_+_79939747 | 0.46 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr3_+_41697046 | 0.43 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr18_+_39126178 | 0.42 |
ENSMUST00000097593.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr5_+_52940391 | 0.42 |
ENSMUST00000031077.13
ENSMUST00000113904.9 ENSMUST00000199840.2 |
Zcchc4
|
zinc finger, CCHC domain containing 4 |
| chr6_-_86710250 | 0.42 |
ENSMUST00000001185.14
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chrM_+_9870 | 0.42 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr17_+_64907697 | 0.42 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chrX_+_139808351 | 0.41 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr5_+_92285748 | 0.41 |
ENSMUST00000031355.10
ENSMUST00000202155.2 |
Uso1
|
USO1 vesicle docking factor |
| chr9_+_117888124 | 0.40 |
ENSMUST00000123690.2
|
Azi2
|
5-azacytidine induced gene 2 |
| chr4_-_87724512 | 0.40 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr3_-_41696906 | 0.40 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr3_+_32490525 | 0.39 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr3_+_60380463 | 0.39 |
ENSMUST00000195077.6
ENSMUST00000193647.6 ENSMUST00000195001.2 ENSMUST00000192807.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr6_-_13607963 | 0.39 |
ENSMUST00000031554.9
ENSMUST00000149123.3 |
Tmem168
|
transmembrane protein 168 |
| chr1_+_167426019 | 0.38 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chrX_+_10118544 | 0.38 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr9_+_78164402 | 0.37 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chrX_-_99638466 | 0.37 |
ENSMUST00000053373.2
|
P2ry4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr3_+_60380243 | 0.36 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr18_-_39000056 | 0.36 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr1_+_13738967 | 0.35 |
ENSMUST00000088542.4
|
Xkr9
|
X-linked Kx blood group related 9 |
| chr6_+_40619913 | 0.35 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chrX_+_10118600 | 0.35 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr2_+_121786892 | 0.35 |
ENSMUST00000110578.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr3_+_32490300 | 0.34 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr3_+_145827410 | 0.34 |
ENSMUST00000039450.5
|
Mcoln3
|
mucolipin 3 |
| chr9_+_66853343 | 0.34 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr11_-_54140462 | 0.34 |
ENSMUST00000019060.6
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr11_-_99213769 | 0.33 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr5_-_151113619 | 0.33 |
ENSMUST00000062015.15
ENSMUST00000110483.9 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr17_+_64203017 | 0.33 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr18_+_39126325 | 0.32 |
ENSMUST00000137497.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr11_-_17903861 | 0.31 |
ENSMUST00000076661.7
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr3_+_66893280 | 0.31 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr13_-_4659120 | 0.31 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr10_-_115198093 | 0.31 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr3_+_66893031 | 0.31 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr12_-_57592907 | 0.31 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr5_-_138617952 | 0.30 |
ENSMUST00000063262.11
ENSMUST00000085852.11 ENSMUST00000110905.9 |
Zfp68
|
zinc finger protein 68 |
| chr3_+_103739877 | 0.29 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr18_-_39620115 | 0.29 |
ENSMUST00000097592.9
ENSMUST00000115571.8 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr1_-_136877277 | 0.28 |
ENSMUST00000168126.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_+_122688721 | 0.28 |
ENSMUST00000023820.6
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr10_+_69761597 | 0.28 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr2_+_121787131 | 0.27 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr6_-_5298455 | 0.27 |
ENSMUST00000057792.9
|
Pon2
|
paraoxonase 2 |
| chr3_+_35808269 | 0.26 |
ENSMUST00000029257.15
|
Atp11b
|
ATPase, class VI, type 11B |
| chr9_-_15212849 | 0.26 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr11_+_21041291 | 0.26 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr9_-_59260713 | 0.25 |
ENSMUST00000026265.8
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr1_+_167425953 | 0.25 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr6_-_83098255 | 0.25 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chr3_-_67422821 | 0.24 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr4_+_85432409 | 0.24 |
ENSMUST00000213098.2
|
Adamtsl1
|
ADAMTS-like 1 |
| chr9_+_53212871 | 0.24 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr10_+_69761314 | 0.24 |
ENSMUST00000182692.8
ENSMUST00000092433.12 |
Ank3
|
ankyrin 3, epithelial |
| chr2_+_91376650 | 0.24 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr12_+_59176543 | 0.24 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr9_+_80072361 | 0.23 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr8_-_41469786 | 0.23 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_-_139908615 | 0.23 |
ENSMUST00000046656.9
ENSMUST00000099304.4 ENSMUST00000110079.9 |
Tasp1
|
taspase, threonine aspartase 1 |
| chr12_-_85335193 | 0.23 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr16_-_30152708 | 0.23 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chr16_-_29363671 | 0.23 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr10_-_115197775 | 0.22 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chrX_+_41157242 | 0.22 |
ENSMUST00000115095.9
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr4_+_74160705 | 0.22 |
ENSMUST00000077851.10
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chrM_+_10167 | 0.22 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr4_+_98919183 | 0.22 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr10_+_69761630 | 0.22 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr8_-_93857899 | 0.22 |
ENSMUST00000034189.17
|
Ces1c
|
carboxylesterase 1C |
| chr4_+_100336003 | 0.22 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr18_+_4375582 | 0.21 |
ENSMUST00000025077.7
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr9_+_118307250 | 0.21 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr17_+_37274714 | 0.20 |
ENSMUST00000209623.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr4_-_32923389 | 0.20 |
ENSMUST00000035719.11
|
Ankrd6
|
ankyrin repeat domain 6 |
| chr4_+_13751297 | 0.20 |
ENSMUST00000105566.9
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr6_+_34723304 | 0.20 |
ENSMUST00000142716.3
|
Cald1
|
caldesmon 1 |
| chr9_+_74883377 | 0.20 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr13_-_3968157 | 0.20 |
ENSMUST00000223258.2
ENSMUST00000091853.12 |
Net1
|
neuroepithelial cell transforming gene 1 |
| chr16_-_38342949 | 0.20 |
ENSMUST00000002925.6
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr10_+_69761784 | 0.20 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr2_-_37249247 | 0.19 |
ENSMUST00000112940.8
ENSMUST00000009174.15 |
Pdcl
|
phosducin-like |
| chr6_+_41512010 | 0.19 |
ENSMUST00000103288.2
|
Trbj1-5
|
T cell receptor beta joining 1-5 |
| chr18_-_57108405 | 0.19 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr11_-_115310743 | 0.19 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr6_-_51443602 | 0.19 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr1_-_155617773 | 0.19 |
ENSMUST00000027740.14
|
Lhx4
|
LIM homeobox protein 4 |
| chr11_-_99313078 | 0.19 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr16_-_44153498 | 0.19 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chrX_+_138511360 | 0.19 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr2_+_124978518 | 0.18 |
ENSMUST00000238754.2
|
Ctxn2
|
cortexin 2 |
| chr7_+_132212349 | 0.18 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr3_-_32546380 | 0.18 |
ENSMUST00000164954.3
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr18_-_66993567 | 0.18 |
ENSMUST00000057942.4
|
Mc4r
|
melanocortin 4 receptor |
| chr11_-_74615496 | 0.18 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr17_+_56312672 | 0.18 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr9_-_96774719 | 0.18 |
ENSMUST00000154146.8
|
Pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr2_+_67935015 | 0.18 |
ENSMUST00000042456.4
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr7_-_110443557 | 0.18 |
ENSMUST00000177462.8
ENSMUST00000176716.3 ENSMUST00000176746.8 ENSMUST00000177236.8 |
Rnf141
|
ring finger protein 141 |
| chr14_-_55822696 | 0.18 |
ENSMUST00000022828.9
|
Emc9
|
ER membrane protein complex subunit 9 |
| chr16_-_44153288 | 0.18 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr14_-_45715308 | 0.17 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr2_-_37537224 | 0.17 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr4_-_32923505 | 0.17 |
ENSMUST00000084749.8
|
Ankrd6
|
ankyrin repeat domain 6 |
| chr2_-_26561294 | 0.17 |
ENSMUST00000028283.2
|
Lcn4
|
lipocalin 4 |
| chr10_+_116111441 | 0.17 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr8_+_106786190 | 0.17 |
ENSMUST00000109308.3
|
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr13_+_102830104 | 0.17 |
ENSMUST00000172138.2
|
Cd180
|
CD180 antigen |
| chr16_+_38167352 | 0.17 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr10_-_81243475 | 0.17 |
ENSMUST00000140916.8
|
Nfic
|
nuclear factor I/C |
| chr17_+_46807637 | 0.17 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr17_-_84154173 | 0.16 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr5_-_88823049 | 0.16 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr17_-_84154196 | 0.16 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr5_+_110088292 | 0.16 |
ENSMUST00000170826.2
|
Gm15446
|
predicted gene 15446 |
| chr18_+_44237474 | 0.16 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr7_+_26456567 | 0.16 |
ENSMUST00000077855.8
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr17_-_37178079 | 0.16 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr14_-_104760051 | 0.16 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr14_+_15369152 | 0.16 |
ENSMUST00000167923.8
|
Gm3696
|
predicted gene 3696 |
| chr7_-_24423715 | 0.16 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr2_-_89671899 | 0.16 |
ENSMUST00000213833.2
|
Olfr1256
|
olfactory receptor 1256 |
| chr2_+_111144362 | 0.16 |
ENSMUST00000219291.2
|
Olfr1280
|
olfactory receptor 1280 |
| chr11_+_43046476 | 0.16 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr5_-_146731810 | 0.16 |
ENSMUST00000085614.6
|
Usp12
|
ubiquitin specific peptidase 12 |
| chr10_-_75946790 | 0.16 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr7_+_28869629 | 0.16 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr14_+_53194239 | 0.15 |
ENSMUST00000199800.2
|
Trav15d-2-dv6d-2
|
T cell receptor alpha variable 15D-2-DV6D-2 |
| chr1_-_80255156 | 0.15 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr16_+_56024676 | 0.15 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr2_+_120807498 | 0.15 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr7_+_28869770 | 0.15 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr4_-_14826587 | 0.15 |
ENSMUST00000117268.9
ENSMUST00000236953.2 |
Otud6b
|
OTU domain containing 6B |
| chr2_+_109721189 | 0.15 |
ENSMUST00000028583.8
|
Lin7c
|
lin-7 homolog C (C. elegans) |
| chr5_+_104350475 | 0.15 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr9_+_105520154 | 0.15 |
ENSMUST00000190358.2
ENSMUST00000191268.7 ENSMUST00000065778.13 ENSMUST00000188784.2 |
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr9_+_21634779 | 0.15 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr4_-_129121676 | 0.15 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr19_-_58781709 | 0.15 |
ENSMUST00000166692.2
|
1700019N19Rik
|
RIKEN cDNA 1700019N19 gene |
| chr18_-_78640066 | 0.15 |
ENSMUST00000235389.2
ENSMUST00000237674.2 |
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr1_-_52992330 | 0.15 |
ENSMUST00000114492.8
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr14_+_52964069 | 0.15 |
ENSMUST00000184883.2
|
Trav6d-3
|
T cell receptor alpha variable 6D-3 |
| chr8_-_123087485 | 0.15 |
ENSMUST00000044123.2
|
Trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr6_+_34722926 | 0.15 |
ENSMUST00000126181.8
|
Cald1
|
caldesmon 1 |
| chr2_-_52448552 | 0.14 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr18_+_38429792 | 0.14 |
ENSMUST00000237211.2
|
Rnf14
|
ring finger protein 14 |
| chr13_+_22508759 | 0.14 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr17_-_78991691 | 0.14 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr1_-_119432455 | 0.14 |
ENSMUST00000004565.15
|
Ralb
|
v-ral simian leukemia viral oncogene B |
| chr3_-_116505469 | 0.14 |
ENSMUST00000153108.6
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr14_+_26300693 | 0.14 |
ENSMUST00000203874.3
ENSMUST00000037585.9 |
Dennd6a
|
DENN/MADD domain containing 6A |
| chrX_+_37689503 | 0.14 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr14_+_53941464 | 0.14 |
ENSMUST00000103664.6
|
Trav5-4
|
T cell receptor alpha variable 5-4 |
| chr17_+_36353844 | 0.14 |
ENSMUST00000097331.2
|
Gm6034
|
predicted gene 6034 |
| chr9_+_65538352 | 0.14 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr1_-_162726053 | 0.13 |
ENSMUST00000143123.3
|
Fmo2
|
flavin containing monooxygenase 2 |
| chr16_+_58548273 | 0.13 |
ENSMUST00000023426.12
ENSMUST00000162057.8 ENSMUST00000162191.2 |
Cldnd1
|
claudin domain containing 1 |
| chr2_+_124978612 | 0.13 |
ENSMUST00000099452.3
ENSMUST00000238377.2 |
Ctxn2
|
cortexin 2 |
| chr7_-_103191924 | 0.13 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612 |
| chr10_+_129907079 | 0.13 |
ENSMUST00000216879.2
|
Olfr822
|
olfactory receptor 822 |
| chrX_-_133012600 | 0.13 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr2_+_61423421 | 0.13 |
ENSMUST00000112495.8
ENSMUST00000112501.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr3_+_66892979 | 0.13 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr2_+_81883566 | 0.13 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr14_-_67309866 | 0.13 |
ENSMUST00000225380.2
ENSMUST00000089230.7 |
Ppp2r2a
|
protein phosphatase 2, regulatory subunit B, alpha |
| chrM_+_7758 | 0.13 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr8_+_21515561 | 0.13 |
ENSMUST00000076754.3
|
Defa21
|
defensin, alpha, 21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prrx1_Isx_Prrxl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.2 | 0.7 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 0.7 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.2 | 1.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.2 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.8 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.3 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.3 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 1.0 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.6 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.2 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.2 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.3 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.3 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 | 0.2 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.2 | GO:2000795 | lung ciliated cell differentiation(GO:0061141) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.3 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:1905167 | regulation of phosphatidylcholine catabolic process(GO:0010899) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.3 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.2 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:2000292 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 1.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 1.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.2 | 0.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 0.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.4 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.4 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.1 | 0.3 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.3 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.7 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.3 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 1.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.6 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.0 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.0 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 1.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |