Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
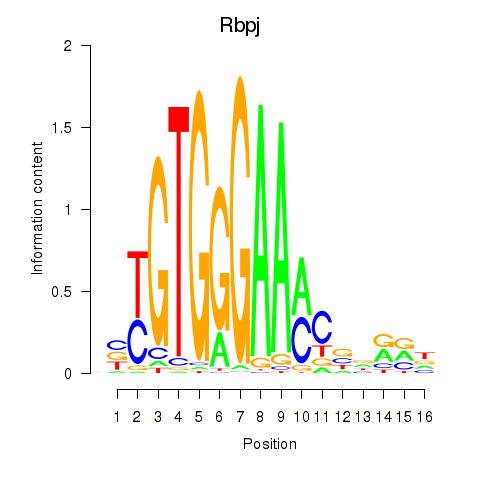
Results for Rbpj
Z-value: 0.87
Transcription factors associated with Rbpj
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rbpj
|
ENSMUSG00000039191.13 | recombination signal binding protein for immunoglobulin kappa J region |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rbpj | mm39_v1_chr5_+_53623458_53623494 | 0.80 | 3.4e-09 | Click! |
Activity profile of Rbpj motif
Sorted Z-values of Rbpj motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_132318039 | 6.33 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr6_+_30639217 | 6.14 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr17_+_37180437 | 5.80 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr7_+_121758646 | 5.74 |
ENSMUST00000033154.8
ENSMUST00000205901.2 |
Plk1
|
polo like kinase 1 |
| chr6_+_29694181 | 5.20 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr14_-_70864448 | 4.90 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr9_+_96140781 | 4.14 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr14_-_20319242 | 4.01 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr18_+_34758062 | 3.72 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr7_+_28240262 | 3.66 |
ENSMUST00000119180.4
|
Sycn
|
syncollin |
| chr11_-_69838971 | 3.44 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr18_+_34757687 | 3.31 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chr5_-_138170644 | 3.31 |
ENSMUST00000000505.16
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr18_+_34757666 | 3.11 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr5_-_138170077 | 3.11 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr14_-_70864666 | 2.98 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr2_+_148640705 | 2.62 |
ENSMUST00000028931.10
ENSMUST00000109947.2 |
Cst8
|
cystatin 8 (cystatin-related epididymal spermatogenic) |
| chr3_-_103716593 | 2.59 |
ENSMUST00000063502.13
ENSMUST00000106832.2 ENSMUST00000106834.8 ENSMUST00000029435.15 |
Dclre1b
|
DNA cross-link repair 1B |
| chr4_-_126096376 | 2.51 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr14_+_75368532 | 2.26 |
ENSMUST00000143539.8
ENSMUST00000134114.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr19_+_53128901 | 2.22 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr9_+_44245981 | 1.99 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr16_+_18247666 | 1.88 |
ENSMUST00000144233.3
|
Txnrd2
|
thioredoxin reductase 2 |
| chr9_+_106699073 | 1.82 |
ENSMUST00000159645.8
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr8_-_89362745 | 1.76 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr1_+_74193138 | 1.76 |
ENSMUST00000027372.8
ENSMUST00000106899.4 |
Cxcr2
|
chemokine (C-X-C motif) receptor 2 |
| chr7_+_29683373 | 1.75 |
ENSMUST00000148442.8
|
Zfp568
|
zinc finger protein 568 |
| chr10_-_93725472 | 1.72 |
ENSMUST00000180375.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr19_+_46044972 | 1.71 |
ENSMUST00000111899.8
ENSMUST00000099392.10 ENSMUST00000062322.11 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chrX_-_73009933 | 1.64 |
ENSMUST00000114372.3
ENSMUST00000033761.13 |
Hcfc1
|
host cell factor C1 |
| chr11_-_80670815 | 1.63 |
ENSMUST00000041065.14
ENSMUST00000070997.6 |
Myo1d
|
myosin ID |
| chr12_-_80306865 | 1.62 |
ENSMUST00000167327.2
|
Actn1
|
actinin, alpha 1 |
| chr19_+_8944369 | 1.58 |
ENSMUST00000052248.8
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr8_+_71047110 | 1.54 |
ENSMUST00000019283.10
ENSMUST00000210005.2 |
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr11_+_63023893 | 1.49 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chr7_-_119058489 | 1.48 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr19_-_10554726 | 1.46 |
ENSMUST00000025568.3
|
Tmem138
|
transmembrane protein 138 |
| chrX_-_36349055 | 1.44 |
ENSMUST00000115231.4
|
Rpl39
|
ribosomal protein L39 |
| chr7_-_6699422 | 1.44 |
ENSMUST00000122432.4
ENSMUST00000002336.16 |
Zim1
|
zinc finger, imprinted 1 |
| chr18_-_34757653 | 1.43 |
ENSMUST00000003876.10
ENSMUST00000115766.8 ENSMUST00000097626.10 ENSMUST00000115765.2 |
Brd8
|
bromodomain containing 8 |
| chr14_+_75368939 | 1.42 |
ENSMUST00000125833.8
ENSMUST00000124499.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr9_+_64718596 | 1.37 |
ENSMUST00000038890.6
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr2_+_164587948 | 1.33 |
ENSMUST00000109327.4
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr13_+_12410240 | 1.29 |
ENSMUST00000059270.10
|
Heatr1
|
HEAT repeat containing 1 |
| chr7_-_99770653 | 1.29 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr19_-_10554417 | 1.27 |
ENSMUST00000236099.2
|
Tmem138
|
transmembrane protein 138 |
| chr19_+_53128861 | 1.25 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr19_-_10554462 | 1.20 |
ENSMUST00000236185.2
|
Tmem138
|
transmembrane protein 138 |
| chrX_+_41238193 | 1.15 |
ENSMUST00000115073.9
ENSMUST00000115072.8 |
Stag2
|
stromal antigen 2 |
| chr7_+_113113061 | 1.13 |
ENSMUST00000129087.8
ENSMUST00000067929.15 ENSMUST00000164745.8 ENSMUST00000136158.8 |
Far1
|
fatty acyl CoA reductase 1 |
| chr7_-_99770280 | 1.11 |
ENSMUST00000208184.2
|
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr17_-_32491339 | 1.05 |
ENSMUST00000237008.2
|
Brd4
|
bromodomain containing 4 |
| chr11_+_63023395 | 1.03 |
ENSMUST00000108701.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr7_+_127566629 | 1.01 |
ENSMUST00000106251.10
ENSMUST00000077609.12 ENSMUST00000121616.9 |
Fus
|
fused in sarcoma |
| chr12_-_80307110 | 0.98 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr2_+_164587901 | 0.98 |
ENSMUST00000017443.14
ENSMUST00000109326.10 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr8_+_111448092 | 0.96 |
ENSMUST00000052457.15
|
Mtss2
|
MTSS I-BAR domain containing 2 |
| chr16_+_21828515 | 0.95 |
ENSMUST00000231632.2
ENSMUST00000232534.2 |
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chr5_+_3593811 | 0.94 |
ENSMUST00000197082.5
ENSMUST00000115527.8 |
Fam133b
|
family with sequence similarity 133, member B |
| chr5_+_136023649 | 0.92 |
ENSMUST00000111142.9
ENSMUST00000111145.10 ENSMUST00000111144.8 ENSMUST00000199239.5 ENSMUST00000005072.10 ENSMUST00000130345.2 |
Dtx2
|
deltex 2, E3 ubiquitin ligase |
| chr2_-_164587836 | 0.92 |
ENSMUST00000109328.8
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr7_+_127376550 | 0.91 |
ENSMUST00000126761.8
ENSMUST00000047157.13 |
Setd1a
|
SET domain containing 1A |
| chr12_+_31315270 | 0.91 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chr2_+_154455217 | 0.88 |
ENSMUST00000081926.13
ENSMUST00000109702.2 |
Zfp341
|
zinc finger protein 341 |
| chr2_+_109111083 | 0.87 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr2_+_154390808 | 0.87 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr4_-_126096551 | 0.86 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr16_-_17540685 | 0.86 |
ENSMUST00000232163.2
ENSMUST00000232202.2 ENSMUST00000080936.14 ENSMUST00000232645.2 ENSMUST00000232431.2 |
Med15
|
mediator complex subunit 15 |
| chr4_-_149251149 | 0.82 |
ENSMUST00000084124.7
|
Pgd
|
phosphogluconate dehydrogenase |
| chr1_+_150268544 | 0.81 |
ENSMUST00000124973.9
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr9_+_64718708 | 0.76 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr19_-_38031774 | 0.74 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr1_-_191307648 | 0.68 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr15_-_54935535 | 0.68 |
ENSMUST00000041733.9
|
Taf2
|
TATA-box binding protein associated factor 2 |
| chr19_+_10554799 | 0.66 |
ENSMUST00000237564.2
ENSMUST00000236743.2 ENSMUST00000235271.2 ENSMUST00000168445.2 ENSMUST00000237641.2 ENSMUST00000236352.2 |
Cyb561a3
|
cytochrome b561 family, member A3 |
| chr16_+_21828223 | 0.65 |
ENSMUST00000023561.8
|
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chr12_-_112640378 | 0.65 |
ENSMUST00000130342.2
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr19_-_46958001 | 0.63 |
ENSMUST00000235234.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr12_+_31315227 | 0.63 |
ENSMUST00000169088.8
|
Lamb1
|
laminin B1 |
| chr5_-_107873883 | 0.63 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr4_-_149747644 | 0.62 |
ENSMUST00000105689.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr7_+_44398064 | 0.59 |
ENSMUST00000165957.8
ENSMUST00000144515.9 |
Vrk3
|
vaccinia related kinase 3 |
| chr19_-_4213347 | 0.58 |
ENSMUST00000025749.15
|
Rps6kb2
|
ribosomal protein S6 kinase, polypeptide 2 |
| chr10_+_40225272 | 0.52 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr1_-_74002156 | 0.52 |
ENSMUST00000191367.2
|
Tns1
|
tensin 1 |
| chr19_+_10554845 | 0.52 |
ENSMUST00000237581.2
|
Cyb561a3
|
cytochrome b561 family, member A3 |
| chr2_-_166904625 | 0.50 |
ENSMUST00000128676.2
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr9_-_114469239 | 0.50 |
ENSMUST00000238809.2
ENSMUST00000216785.2 |
Cnot10
|
CCR4-NOT transcription complex, subunit 10 |
| chr1_+_87254729 | 0.49 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr11_+_78215026 | 0.47 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr4_-_117772163 | 0.43 |
ENSMUST00000036156.6
|
Ipo13
|
importin 13 |
| chr1_+_150269001 | 0.42 |
ENSMUST00000119161.9
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr13_-_67632333 | 0.42 |
ENSMUST00000223868.2
ENSMUST00000019572.9 |
Zfp874b
|
zinc finger protein 874b |
| chr7_-_79765042 | 0.41 |
ENSMUST00000206714.2
ENSMUST00000107384.10 |
Idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr9_+_35470752 | 0.41 |
ENSMUST00000034615.10
ENSMUST00000121246.2 |
Pus3
|
pseudouridine synthase 3 |
| chr16_-_17540805 | 0.39 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr19_+_10554510 | 0.38 |
ENSMUST00000237814.2
|
Cyb561a3
|
cytochrome b561 family, member A3 |
| chr7_-_105401398 | 0.38 |
ENSMUST00000033184.6
|
Tpp1
|
tripeptidyl peptidase I |
| chr16_+_20470402 | 0.38 |
ENSMUST00000007212.9
ENSMUST00000232629.2 |
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr7_+_44398008 | 0.37 |
ENSMUST00000171821.8
|
Vrk3
|
vaccinia related kinase 3 |
| chr5_-_88823049 | 0.37 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chrX_+_98179725 | 0.37 |
ENSMUST00000052839.7
|
Efnb1
|
ephrin B1 |
| chr19_+_46045675 | 0.36 |
ENSMUST00000126127.8
ENSMUST00000147640.2 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr5_+_115465205 | 0.35 |
ENSMUST00000031513.14
|
Srsf9
|
serine and arginine-rich splicing factor 9 |
| chr2_+_120439858 | 0.35 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr1_+_74317709 | 0.32 |
ENSMUST00000077985.4
|
Gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr11_-_62348599 | 0.32 |
ENSMUST00000127471.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr2_+_112097087 | 0.32 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr16_-_89940652 | 0.31 |
ENSMUST00000114124.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr9_-_114469121 | 0.30 |
ENSMUST00000070117.8
|
Cnot10
|
CCR4-NOT transcription complex, subunit 10 |
| chrX_-_8118541 | 0.30 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr7_+_5023552 | 0.29 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr19_+_41921903 | 0.28 |
ENSMUST00000224258.2
ENSMUST00000026154.9 ENSMUST00000224896.2 |
Zdhhc16
|
zinc finger, DHHC domain containing 16 |
| chr7_+_45289391 | 0.28 |
ENSMUST00000148532.4
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr8_+_120588977 | 0.27 |
ENSMUST00000034287.10
|
Klhl36
|
kelch-like 36 |
| chr1_+_171386752 | 0.27 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr3_-_89177796 | 0.26 |
ENSMUST00000107460.8
ENSMUST00000029565.11 ENSMUST00000130230.2 |
Slc50a1
|
solute carrier family 50 (sugar transporter), member 1 |
| chr19_+_36061096 | 0.25 |
ENSMUST00000025714.9
|
Rpp30
|
ribonuclease P/MRP 30 subunit |
| chr6_-_5298455 | 0.25 |
ENSMUST00000057792.9
|
Pon2
|
paraoxonase 2 |
| chr7_+_78922947 | 0.23 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr1_+_21021889 | 0.23 |
ENSMUST00000038447.6
|
Efhc1
|
EF-hand domain (C-terminal) containing 1 |
| chr8_-_22550321 | 0.22 |
ENSMUST00000006742.11
|
Atp7b
|
ATPase, Cu++ transporting, beta polypeptide |
| chr18_+_37880027 | 0.19 |
ENSMUST00000193404.2
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr3_+_103716836 | 0.18 |
ENSMUST00000076599.8
ENSMUST00000106824.8 ENSMUST00000106823.8 ENSMUST00000047285.7 |
Ap4b1
|
adaptor-related protein complex AP-4, beta 1 |
| chr8_+_71047010 | 0.18 |
ENSMUST00000211117.2
|
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr11_-_62348115 | 0.18 |
ENSMUST00000069456.11
ENSMUST00000018645.13 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr15_+_83447784 | 0.18 |
ENSMUST00000047419.8
|
Tspo
|
translocator protein |
| chr2_-_120439981 | 0.16 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr4_-_126861918 | 0.16 |
ENSMUST00000106108.9
|
Zmym4
|
zinc finger, MYM-type 4 |
| chr2_+_26471062 | 0.15 |
ENSMUST00000238951.2
ENSMUST00000166920.10 |
Egfl7
|
EGF-like domain 7 |
| chr2_-_180883813 | 0.14 |
ENSMUST00000094203.11
ENSMUST00000108831.8 |
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr14_-_54647647 | 0.13 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr7_+_113113037 | 0.13 |
ENSMUST00000033018.15
|
Far1
|
fatty acyl CoA reductase 1 |
| chr7_+_111825063 | 0.13 |
ENSMUST00000050149.12
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr16_+_32882882 | 0.13 |
ENSMUST00000023497.3
|
Lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr12_-_115459678 | 0.13 |
ENSMUST00000103534.2
|
Ighv1-63
|
immunoglobulin heavy variable V1-63 |
| chr8_-_105368298 | 0.12 |
ENSMUST00000093234.5
|
Ciao2b
|
cytosolic iron-sulfur assembly component 2B |
| chr5_-_76139107 | 0.12 |
ENSMUST00000113516.2
|
Kdr
|
kinase insert domain protein receptor |
| chr5_+_138185747 | 0.11 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr12_-_14202041 | 0.11 |
ENSMUST00000020926.8
|
Lratd1
|
LRAT domain containing 1 |
| chr18_+_63111005 | 0.10 |
ENSMUST00000235372.2
ENSMUST00000237483.2 |
Napg
|
N-ethylmaleimide sensitive fusion protein attachment protein gamma |
| chr2_-_109111064 | 0.09 |
ENSMUST00000147770.2
|
Mettl15
|
methyltransferase like 15 |
| chr14_-_49304110 | 0.08 |
ENSMUST00000162175.9
|
Exoc5
|
exocyst complex component 5 |
| chr7_-_44397730 | 0.08 |
ENSMUST00000118162.8
ENSMUST00000140599.9 ENSMUST00000120798.8 |
Zfp473
|
zinc finger protein 473 |
| chr7_-_24937276 | 0.08 |
ENSMUST00000071739.12
ENSMUST00000108411.2 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr11_+_69871952 | 0.08 |
ENSMUST00000108593.8
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr11_+_76297969 | 0.07 |
ENSMUST00000021203.7
ENSMUST00000152183.2 |
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr7_+_44397837 | 0.07 |
ENSMUST00000002275.15
|
Vrk3
|
vaccinia related kinase 3 |
| chr11_-_6576030 | 0.07 |
ENSMUST00000000394.14
ENSMUST00000189268.7 ENSMUST00000136682.8 |
Tbrg4
|
transforming growth factor beta regulated gene 4 |
| chr18_+_37794819 | 0.07 |
ENSMUST00000194888.2
ENSMUST00000194190.2 |
Pcdhga1
|
protocadherin gamma subfamily A, 1 |
| chr2_+_104961228 | 0.07 |
ENSMUST00000111098.8
ENSMUST00000111099.2 |
Wt1
|
Wilms tumor 1 homolog |
| chr16_+_44764046 | 0.06 |
ENSMUST00000164007.8
ENSMUST00000171779.8 |
Cd200r3
|
CD200 receptor 3 |
| chr11_+_109376432 | 0.06 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr16_+_11140779 | 0.06 |
ENSMUST00000115814.4
|
Snx29
|
sorting nexin 29 |
| chr2_+_59442378 | 0.06 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr18_+_37869950 | 0.05 |
ENSMUST00000091935.7
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr2_+_128705829 | 0.05 |
ENSMUST00000028864.3
|
Fbln7
|
fibulin 7 |
| chr8_+_95744320 | 0.05 |
ENSMUST00000051259.10
|
Adgrg3
|
adhesion G protein-coupled receptor G3 |
| chr11_-_69872050 | 0.04 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr3_-_87792865 | 0.04 |
ENSMUST00000005015.10
|
Prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr11_+_54205722 | 0.04 |
ENSMUST00000072178.11
ENSMUST00000101211.9 ENSMUST00000101213.9 ENSMUST00000064690.10 ENSMUST00000108899.8 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr2_-_120439826 | 0.04 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr12_+_33365371 | 0.03 |
ENSMUST00000154742.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr12_+_83734919 | 0.03 |
ENSMUST00000041806.13
|
Psen1
|
presenilin 1 |
| chr7_-_29750785 | 0.02 |
ENSMUST00000207072.2
ENSMUST00000207873.2 |
Zfp14
|
zinc finger protein 14 |
| chr7_-_10229249 | 0.02 |
ENSMUST00000032551.8
|
Zik1
|
zinc finger protein interacting with K protein 1 |
| chr5_-_24782465 | 0.02 |
ENSMUST00000030795.10
|
Abcf2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr9_-_95632387 | 0.01 |
ENSMUST00000189137.7
ENSMUST00000053785.10 |
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr2_+_83474779 | 0.01 |
ENSMUST00000081591.7
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rbpj
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 1.3 | 7.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.6 | 2.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.6 | 1.8 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.6 | 1.7 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.5 | 6.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 1.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.4 | 10.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.4 | 1.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.4 | 1.2 | GO:0046832 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.4 | 1.6 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.4 | 1.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 1.8 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.3 | 2.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 | 5.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.2 | 6.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 4.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.8 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 | 2.7 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) |
| 0.2 | 0.6 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 0.6 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 1.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.4 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.1 | 0.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 1.7 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 3.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 0.3 | GO:1904266 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.1 | 1.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.3 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 2.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.6 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 2.5 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 1.1 | GO:0044154 | histone H4-K12 acetylation(GO:0043983) histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 4.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.3 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 1.9 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 1.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 1.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:2001074 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.0 | 0.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.4 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.4 | GO:0031295 | T cell costimulation(GO:0031295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.1 | 7.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.6 | 2.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.5 | 1.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.4 | 5.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.4 | 1.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.4 | 6.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 3.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 2.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 11.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 3.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 4.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 2.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 2.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.9 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 3.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.7 | 3.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.6 | 1.8 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.6 | 1.7 | GO:0016872 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.4 | 1.6 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 2.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 0.8 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 0.7 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 4.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 1.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.2 | 6.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.9 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.2 | 1.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 6.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.6 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 10.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 4.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 7.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.3 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 1.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.3 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 1.7 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 2.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 3.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 2.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 17.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 6.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 11.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 5.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.3 | 4.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 2.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 3.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |