Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
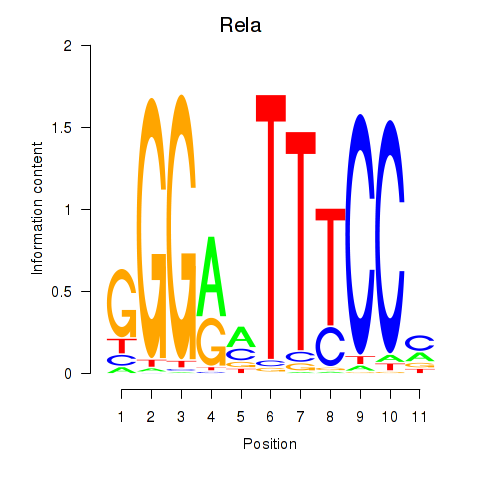
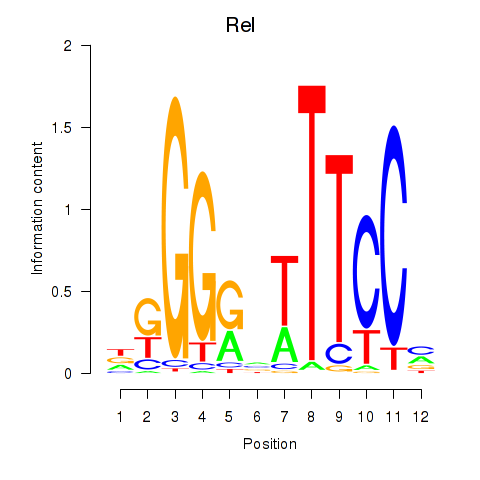
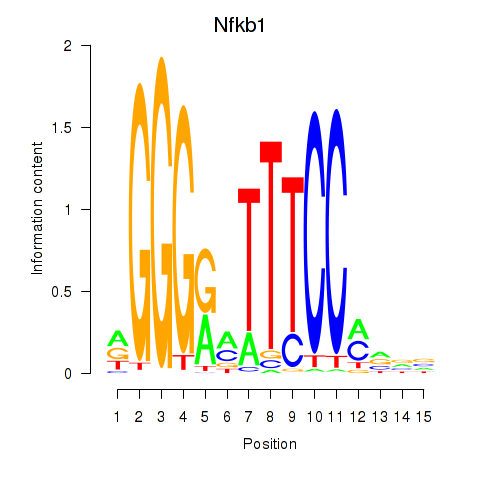
Results for Rela_Rel_Nfkb1
Z-value: 1.31
Transcription factors associated with Rela_Rel_Nfkb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rela
|
ENSMUSG00000024927.9 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) |
|
Rel
|
ENSMUSG00000020275.10 | reticuloendotheliosis oncogene |
|
Nfkb1
|
ENSMUSG00000028163.18 | nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb1 | mm39_v1_chr3_-_135373560_135373582 | 0.61 | 7.1e-05 | Click! |
| Rel | mm39_v1_chr11_-_23720953_23721028 | 0.50 | 1.9e-03 | Click! |
| Rela | mm39_v1_chr19_+_5687503_5687514 | 0.41 | 1.4e-02 | Click! |
Activity profile of Rela_Rel_Nfkb1 motif
Sorted Z-values of Rela_Rel_Nfkb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_92496730 | 4.51 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr11_-_83421333 | 4.34 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr18_+_60936910 | 4.19 |
ENSMUST00000097563.9
ENSMUST00000050487.16 ENSMUST00000167610.2 |
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) |
| chr17_-_28736483 | 3.82 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr17_+_43978377 | 3.53 |
ENSMUST00000233627.2
ENSMUST00000233437.2 |
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr17_+_43978280 | 3.43 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr17_+_37504783 | 3.34 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr7_+_25872836 | 3.19 |
ENSMUST00000082214.5
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr13_+_112600604 | 2.91 |
ENSMUST00000183663.8
ENSMUST00000184311.8 ENSMUST00000183886.8 |
Il6st
|
interleukin 6 signal transducer |
| chr4_+_131649001 | 2.67 |
ENSMUST00000094666.4
|
Tmem200b
|
transmembrane protein 200B |
| chr16_+_31241085 | 2.47 |
ENSMUST00000089759.9
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr8_-_11329656 | 2.45 |
ENSMUST00000208095.2
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr5_+_91039092 | 2.42 |
ENSMUST00000031327.9
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr9_-_14526246 | 2.37 |
ENSMUST00000013220.8
|
Amotl1
|
angiomotin-like 1 |
| chr11_-_99383938 | 2.30 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr9_-_70841881 | 2.22 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr19_-_7943365 | 2.19 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr3_+_104696108 | 2.18 |
ENSMUST00000002303.12
|
Rhoc
|
ras homolog family member C |
| chr10_+_111342147 | 2.18 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1 |
| chr7_-_142223662 | 2.13 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr10_-_95251327 | 2.04 |
ENSMUST00000172070.8
ENSMUST00000150432.8 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr7_-_99276310 | 1.98 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr17_-_35081456 | 1.97 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr8_-_62355690 | 1.96 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr7_-_25488060 | 1.94 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr18_-_35855383 | 1.90 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr10_-_95251145 | 1.89 |
ENSMUST00000119917.2
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr10_-_127843377 | 1.86 |
ENSMUST00000219447.2
ENSMUST00000219780.2 ENSMUST00000219707.2 ENSMUST00000219953.2 ENSMUST00000219183.2 |
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr11_-_48884999 | 1.84 |
ENSMUST00000146439.8
|
Tgtp1
|
T cell specific GTPase 1 |
| chr7_-_19363280 | 1.77 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr8_+_72128920 | 1.73 |
ENSMUST00000110013.10
ENSMUST00000051995.14 |
Jak3
|
Janus kinase 3 |
| chr9_-_70842090 | 1.71 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr5_-_105441554 | 1.66 |
ENSMUST00000050011.10
ENSMUST00000196520.2 |
Gm43302
Gbp6
|
predicted gene 43302 guanylate binding protein 6 |
| chr12_+_111383864 | 1.63 |
ENSMUST00000220537.2
ENSMUST00000223050.2 ENSMUST00000072646.8 ENSMUST00000223431.2 ENSMUST00000221144.2 ENSMUST00000222437.2 |
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr17_-_45997823 | 1.62 |
ENSMUST00000156254.8
|
Tmem63b
|
transmembrane protein 63b |
| chr9_-_44646487 | 1.58 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr14_+_33662976 | 1.57 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr12_-_35584968 | 1.55 |
ENSMUST00000116436.9
|
Ahr
|
aryl-hydrocarbon receptor |
| chr4_+_137408975 | 1.54 |
ENSMUST00000047243.12
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr17_-_56312555 | 1.52 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr7_+_75498058 | 1.51 |
ENSMUST00000171155.4
ENSMUST00000092073.11 ENSMUST00000206019.2 ENSMUST00000205612.2 ENSMUST00000205887.2 |
Klhl25
|
kelch-like 25 |
| chrX_-_73290140 | 1.51 |
ENSMUST00000101454.9
ENSMUST00000033699.13 |
Flna
|
filamin, alpha |
| chr19_+_3372296 | 1.47 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr7_-_44465043 | 1.47 |
ENSMUST00000107893.9
|
Atf5
|
activating transcription factor 5 |
| chr1_+_172327569 | 1.46 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr13_+_43938251 | 1.44 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chrX_-_73289970 | 1.42 |
ENSMUST00000130007.8
|
Flna
|
filamin, alpha |
| chr11_+_101066867 | 1.39 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr17_+_35598583 | 1.39 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr12_+_104180795 | 1.32 |
ENSMUST00000121337.8
ENSMUST00000167049.8 ENSMUST00000101080.2 |
Serpina3f
|
serine (or cysteine) peptidase inhibitor, clade A, member 3F |
| chr1_+_172327812 | 1.30 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr3_+_96508400 | 1.29 |
ENSMUST00000062058.5
|
Lix1l
|
Lix1-like |
| chr14_-_45767421 | 1.28 |
ENSMUST00000150660.3
|
Fermt2
|
fermitin family member 2 |
| chr4_-_101122433 | 1.27 |
ENSMUST00000149297.2
ENSMUST00000102781.10 |
Jak1
|
Janus kinase 1 |
| chr9_+_32607301 | 1.25 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr17_-_34506744 | 1.22 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr10_-_127587576 | 1.22 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr18_+_11972277 | 1.22 |
ENSMUST00000171109.9
ENSMUST00000046948.10 |
Cables1
|
CDK5 and Abl enzyme substrate 1 |
| chr16_+_44913974 | 1.21 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr2_-_32278245 | 1.21 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr1_+_87983099 | 1.19 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr5_+_8010445 | 1.18 |
ENSMUST00000115421.3
|
Steap4
|
STEAP family member 4 |
| chr17_+_35413415 | 1.16 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr2_-_32277773 | 1.16 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr1_+_134121170 | 1.15 |
ENSMUST00000038445.13
ENSMUST00000191577.2 |
Mybph
|
myosin binding protein H |
| chr3_-_83749036 | 1.14 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chr1_+_171041539 | 1.14 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr17_+_56259617 | 1.12 |
ENSMUST00000003274.8
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr14_-_70585874 | 1.12 |
ENSMUST00000152067.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr12_+_29988035 | 1.11 |
ENSMUST00000122328.8
ENSMUST00000118321.3 |
Pxdn
|
peroxidasin |
| chr12_-_80307110 | 1.11 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr18_-_20192535 | 1.10 |
ENSMUST00000075214.9
ENSMUST00000039247.11 |
Dsc2
|
desmocollin 2 |
| chr4_-_107164315 | 1.09 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr1_+_171041583 | 1.08 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr6_+_48818410 | 1.07 |
ENSMUST00000101426.11
ENSMUST00000168406.4 |
Tmem176a
|
transmembrane protein 176A |
| chr11_+_70431063 | 1.06 |
ENSMUST00000018429.12
ENSMUST00000108557.10 ENSMUST00000108556.2 |
Pld2
|
phospholipase D2 |
| chr12_+_84085709 | 1.04 |
ENSMUST00000221229.2
|
Gm49366
|
predicted gene, 49366 |
| chr6_-_41012435 | 1.01 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr16_-_18052937 | 1.01 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr17_-_24424456 | 1.00 |
ENSMUST00000201583.2
ENSMUST00000202925.4 ENSMUST00000167791.9 ENSMUST00000201960.4 ENSMUST00000040474.11 ENSMUST00000201089.4 ENSMUST00000201301.4 ENSMUST00000201805.4 ENSMUST00000168410.9 ENSMUST00000097376.10 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr10_-_86843878 | 1.00 |
ENSMUST00000035288.17
|
Stab2
|
stabilin 2 |
| chr1_-_172418058 | 1.00 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr1_+_87983189 | 0.96 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr11_-_75329726 | 0.95 |
ENSMUST00000108437.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr9_+_90045109 | 0.94 |
ENSMUST00000113059.8
ENSMUST00000167122.8 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr11_-_54846873 | 0.94 |
ENSMUST00000155316.2
ENSMUST00000108889.10 ENSMUST00000126703.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr4_+_131648509 | 0.93 |
ENSMUST00000238733.2
|
Tmem200b
|
transmembrane protein 200B |
| chr16_-_17711950 | 0.93 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr8_+_89309408 | 0.93 |
ENSMUST00000211113.2
|
Nkd1
|
naked cuticle 1 |
| chr16_+_32734464 | 0.92 |
ENSMUST00000023491.13
ENSMUST00000170899.8 ENSMUST00000170201.8 ENSMUST00000165616.8 ENSMUST00000135193.9 |
Lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr17_+_25097199 | 0.91 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr4_+_136013372 | 0.91 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr1_-_135186176 | 0.91 |
ENSMUST00000185752.2
ENSMUST00000003135.14 |
Elf3
|
E74-like factor 3 |
| chr14_-_104081119 | 0.90 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr19_+_34268053 | 0.90 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr18_+_36693646 | 0.89 |
ENSMUST00000155329.9
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr2_-_118377500 | 0.88 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor |
| chr9_+_107784065 | 0.87 |
ENSMUST00000035203.9
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr10_+_128139227 | 0.86 |
ENSMUST00000218315.2
ENSMUST00000219721.2 |
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr2_+_91090167 | 0.86 |
ENSMUST00000138470.2
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr11_-_106050724 | 0.86 |
ENSMUST00000064545.11
|
Limd2
|
LIM domain containing 2 |
| chr5_-_105287405 | 0.85 |
ENSMUST00000100961.5
ENSMUST00000031235.13 ENSMUST00000197799.2 ENSMUST00000199629.2 ENSMUST00000196677.5 ENSMUST00000100962.8 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr11_-_69771797 | 0.83 |
ENSMUST00000238978.2
|
Kctd11
|
potassium channel tetramerisation domain containing 11 |
| chr14_-_45767575 | 0.81 |
ENSMUST00000045905.15
|
Fermt2
|
fermitin family member 2 |
| chr2_-_6217844 | 0.80 |
ENSMUST00000042658.5
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3 |
| chr2_-_5068807 | 0.80 |
ENSMUST00000114996.8
|
Optn
|
optineurin |
| chr12_+_86725459 | 0.78 |
ENSMUST00000021681.4
|
Vash1
|
vasohibin 1 |
| chr2_-_32321116 | 0.77 |
ENSMUST00000127961.3
ENSMUST00000136361.8 ENSMUST00000052119.14 |
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr3_-_135397298 | 0.76 |
ENSMUST00000029812.14
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr3_+_142326363 | 0.75 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2 |
| chr11_-_30148230 | 0.75 |
ENSMUST00000102838.10
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr19_+_34268071 | 0.75 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr14_-_70588803 | 0.75 |
ENSMUST00000143153.2
ENSMUST00000127000.2 ENSMUST00000068044.14 ENSMUST00000022688.10 |
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr2_-_160155536 | 0.74 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chr2_+_13578738 | 0.74 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr6_-_48818430 | 0.73 |
ENSMUST00000205147.3
|
Tmem176b
|
transmembrane protein 176B |
| chr11_-_54853729 | 0.72 |
ENSMUST00000108885.8
ENSMUST00000102730.9 ENSMUST00000018482.13 ENSMUST00000108886.8 ENSMUST00000102731.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr11_-_51647204 | 0.71 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr14_+_14475188 | 0.71 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr7_+_95860863 | 0.71 |
ENSMUST00000107165.8
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr6_-_48818006 | 0.71 |
ENSMUST00000203229.3
|
Tmem176b
|
transmembrane protein 176B |
| chr11_+_100750316 | 0.71 |
ENSMUST00000107356.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr15_+_6599001 | 0.70 |
ENSMUST00000227175.2
|
Fyb
|
FYN binding protein |
| chr5_+_72805153 | 0.70 |
ENSMUST00000197837.3
|
Nipal1
|
NIPA-like domain containing 1 |
| chr5_-_134707042 | 0.70 |
ENSMUST00000111233.8
|
Limk1
|
LIM-domain containing, protein kinase |
| chr1_-_60606237 | 0.70 |
ENSMUST00000142258.3
|
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr3_-_121056944 | 0.70 |
ENSMUST00000128909.8
ENSMUST00000029777.14 |
Tlcd4
|
TLC domain containing 4 |
| chr12_-_101785307 | 0.70 |
ENSMUST00000021603.9
|
Fbln5
|
fibulin 5 |
| chr16_+_23109213 | 0.70 |
ENSMUST00000115335.2
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr2_-_5068743 | 0.69 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chr11_+_69471185 | 0.69 |
ENSMUST00000171247.8
ENSMUST00000108658.10 ENSMUST00000005371.12 |
Trp53
|
transformation related protein 53 |
| chr3_+_37694094 | 0.69 |
ENSMUST00000108109.8
ENSMUST00000038569.8 ENSMUST00000108107.2 |
Spry1
|
sprouty RTK signaling antagonist 1 |
| chr6_-_48818302 | 0.69 |
ENSMUST00000203355.3
ENSMUST00000166247.8 |
Tmem176b
|
transmembrane protein 176B |
| chr8_-_25591737 | 0.69 |
ENSMUST00000098866.11
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr10_+_128626772 | 0.68 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr7_-_109215960 | 0.68 |
ENSMUST00000077909.9
|
Denn2b
|
DENN domain containing 2B |
| chr16_-_30086317 | 0.67 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr11_+_48977852 | 0.67 |
ENSMUST00000046704.7
ENSMUST00000203810.3 ENSMUST00000203149.3 |
Ifi47
Olfr56
|
interferon gamma inducible protein 47 olfactory receptor 56 |
| chr1_-_36312482 | 0.66 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr13_-_36918424 | 0.66 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
| chr7_+_28466160 | 0.66 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr3_-_115923098 | 0.66 |
ENSMUST00000196449.5
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr5_-_38649291 | 0.65 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr2_-_166902307 | 0.65 |
ENSMUST00000155281.8
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr13_+_93810911 | 0.65 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr10_+_128139191 | 0.65 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr12_-_80306865 | 0.65 |
ENSMUST00000167327.2
|
Actn1
|
actinin, alpha 1 |
| chr4_+_130643260 | 0.65 |
ENSMUST00000030316.7
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr8_-_25592001 | 0.64 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr11_+_104441489 | 0.64 |
ENSMUST00000018800.9
|
Myl4
|
myosin, light polypeptide 4 |
| chr11_-_83469446 | 0.64 |
ENSMUST00000019266.6
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr2_+_84880776 | 0.64 |
ENSMUST00000111605.9
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr6_-_48817914 | 0.63 |
ENSMUST00000164733.4
|
Tmem176b
|
transmembrane protein 176B |
| chr11_+_48977888 | 0.63 |
ENSMUST00000214804.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr3_-_107667499 | 0.63 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr6_+_15720653 | 0.63 |
ENSMUST00000101663.10
ENSMUST00000190255.7 ENSMUST00000189359.7 ENSMUST00000125326.8 |
Mdfic
|
MyoD family inhibitor domain containing |
| chr15_-_89263128 | 0.63 |
ENSMUST00000227834.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr3_+_89987749 | 0.63 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr10_-_81266800 | 0.62 |
ENSMUST00000117966.2
|
Nfic
|
nuclear factor I/C |
| chr8_-_25592385 | 0.61 |
ENSMUST00000064883.14
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chrX_+_135039745 | 0.61 |
ENSMUST00000116527.2
|
Bex4
|
brain expressed X-linked 4 |
| chr11_+_97690585 | 0.61 |
ENSMUST00000129558.8
|
Lasp1
|
LIM and SH3 protein 1 |
| chr3_+_142202642 | 0.61 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr1_-_173707677 | 0.60 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chr9_-_14526768 | 0.59 |
ENSMUST00000162901.8
|
Amotl1
|
angiomotin-like 1 |
| chr19_-_10582773 | 0.59 |
ENSMUST00000237788.2
|
Tkfc
|
triokinase, FMN cyclase |
| chr9_+_69361348 | 0.59 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr14_-_54655079 | 0.58 |
ENSMUST00000226753.2
ENSMUST00000197440.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr19_-_10582672 | 0.58 |
ENSMUST00000236478.2
ENSMUST00000236950.2 |
Tkfc
|
triokinase, FMN cyclase |
| chr17_-_45996899 | 0.57 |
ENSMUST00000145873.8
|
Tmem63b
|
transmembrane protein 63b |
| chr17_-_45997046 | 0.57 |
ENSMUST00000143907.3
ENSMUST00000127065.8 |
Tmem63b
|
transmembrane protein 63b |
| chr1_-_170886924 | 0.57 |
ENSMUST00000164044.8
ENSMUST00000169017.8 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr3_-_67371161 | 0.57 |
ENSMUST00000058981.3
|
Lxn
|
latexin |
| chr2_+_29855572 | 0.57 |
ENSMUST00000113719.9
ENSMUST00000113717.8 ENSMUST00000113741.8 ENSMUST00000100225.9 ENSMUST00000095083.11 ENSMUST00000046257.14 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr11_+_97689819 | 0.57 |
ENSMUST00000143571.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr15_+_81629258 | 0.57 |
ENSMUST00000109554.3
ENSMUST00000230946.2 |
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr6_-_48818044 | 0.57 |
ENSMUST00000101429.11
ENSMUST00000204073.3 |
Tmem176b
|
transmembrane protein 176B |
| chr17_+_34406523 | 0.56 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr4_+_102446883 | 0.56 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr7_-_44180700 | 0.56 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr19_-_58442866 | 0.56 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr2_+_13579092 | 0.56 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr18_-_4352944 | 0.54 |
ENSMUST00000025078.10
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr14_-_36690636 | 0.54 |
ENSMUST00000067700.13
|
Ccser2
|
coiled-coil serine rich 2 |
| chr13_-_64518112 | 0.54 |
ENSMUST00000021933.8
ENSMUST00000222462.2 |
Ctsl
|
cathepsin L |
| chr5_+_65127412 | 0.53 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr13_+_31740117 | 0.53 |
ENSMUST00000042118.11
|
Foxq1
|
forkhead box Q1 |
| chr18_+_84869456 | 0.53 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr6_+_68233361 | 0.53 |
ENSMUST00000103320.3
|
Igkv14-111
|
immunoglobulin kappa variable 14-111 |
| chr9_-_7873171 | 0.52 |
ENSMUST00000159323.2
ENSMUST00000115673.3 |
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr11_+_100751272 | 0.52 |
ENSMUST00000107357.4
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr6_-_137146708 | 0.52 |
ENSMUST00000117919.8
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr9_+_69360902 | 0.52 |
ENSMUST00000034756.15
ENSMUST00000123470.8 |
Anxa2
|
annexin A2 |
| chr2_-_164285097 | 0.52 |
ENSMUST00000017153.4
|
Sdc4
|
syndecan 4 |
| chr6_-_125208738 | 0.52 |
ENSMUST00000043422.8
|
Tapbpl
|
TAP binding protein-like |
| chr4_-_45012093 | 0.52 |
ENSMUST00000131991.2
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr12_+_111132908 | 0.51 |
ENSMUST00000139162.8
ENSMUST00000060274.7 |
Traf3
|
TNF receptor-associated factor 3 |
| chr17_-_45997132 | 0.51 |
ENSMUST00000113523.9
|
Tmem63b
|
transmembrane protein 63b |
| chr18_+_4994600 | 0.51 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
| chr17_+_34406762 | 0.51 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rela_Rel_Nfkb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 1.1 | 3.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 1.1 | 4.5 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 1.1 | 4.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.0 | 2.9 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.9 | 2.8 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.8 | 4.2 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.8 | 2.4 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.8 | 2.4 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.8 | 3.9 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.7 | 3.7 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.5 | 1.6 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.5 | 1.4 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.4 | 1.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 1.1 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) detection of triacyl bacterial lipopeptide(GO:0042495) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.4 | 1.1 | GO:1904024 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.4 | 4.9 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.4 | 1.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.3 | 1.0 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.3 | 0.6 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.3 | 1.7 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 1.1 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.3 | 2.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 | 0.8 | GO:1990051 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.3 | 1.5 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.3 | 3.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 1.3 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.3 | 0.8 | GO:1904632 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.2 | 1.5 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.2 | 1.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.7 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 0.6 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.2 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 0.8 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 0.6 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.2 | 1.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.2 | 2.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.7 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.2 | 0.4 | GO:0046226 | coumarin catabolic process(GO:0046226) |
| 0.2 | 0.5 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.2 | 0.7 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 | 0.7 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.2 | 1.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.2 | 1.3 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 0.8 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 0.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 1.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 4.2 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.9 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 2.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.4 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.4 | GO:0042668 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.1 | 1.4 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 | 0.3 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.7 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.1 | 1.2 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 3.4 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.9 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 1.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.5 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 1.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 3.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 1.0 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.1 | 1.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 2.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.5 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.4 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 1.5 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 3.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 2.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 1.5 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.3 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 0.8 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 0.8 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 1.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 1.5 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 0.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 2.0 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 1.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 2.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.7 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.6 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.1 | 0.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 1.5 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.3 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.2 | GO:0034148 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.4 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 1.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 1.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.4 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.2 | GO:0015744 | succinate transport(GO:0015744) |
| 0.1 | 0.3 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.0 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 1.7 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 1.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.7 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.2 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.5 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 0.9 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.7 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.4 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.5 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.5 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.7 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.3 | GO:0032423 | regulation of mismatch repair(GO:0032423) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.3 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.9 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.4 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) mast cell migration(GO:0097531) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 1.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 1.0 | GO:0050951 | sensory perception of temperature stimulus(GO:0050951) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 2.0 | GO:0060193 | positive regulation of lipase activity(GO:0060193) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.4 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.7 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.9 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0002856 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.6 | GO:0032743 | positive regulation of interleukin-2 production(GO:0032743) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.6 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.3 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.3 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.9 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 1.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.0 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 1.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.2 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.5 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.1 | GO:0002692 | negative regulation of cellular extravasation(GO:0002692) negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0034143 | regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 1.0 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.0 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.5 | 2.9 | GO:0031523 | Myb complex(GO:0031523) |
| 0.5 | 5.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 1.8 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.4 | 1.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.4 | 1.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.3 | 2.4 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 1.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 1.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 3.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 3.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 2.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 1.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 1.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 0.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.9 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 4.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.9 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 2.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 3.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.6 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 2.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 3.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 1.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 3.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 1.4 | 4.3 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 1.0 | 2.9 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.8 | 5.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.8 | 2.5 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.8 | 3.9 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.7 | 4.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.7 | 3.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 1.5 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.5 | 2.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.4 | 1.6 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.4 | 1.2 | GO:0050354 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.4 | 1.1 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.4 | 1.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.3 | 0.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 0.9 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 1.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 1.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 0.7 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 1.9 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 1.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 0.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 3.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 2.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 2.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 1.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 0.7 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 2.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 0.4 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 1.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 1.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 1.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.2 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.5 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 1.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.5 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.1 | 0.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 3.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 2.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.6 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.4 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.1 | 3.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.3 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 2.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.6 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 2.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 5.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 3.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 4.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 6.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 2.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 3.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 4.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 3.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 1.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 4.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 8.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 3.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 5.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 2.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 3.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |