Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
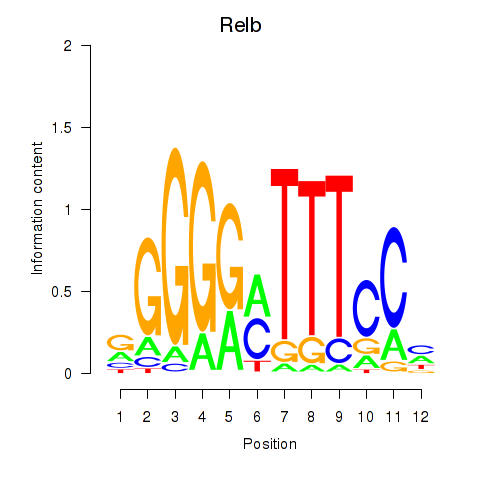
Results for Relb
Z-value: 0.41
Transcription factors associated with Relb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Relb
|
ENSMUSG00000002983.18 | avian reticuloendotheliosis viral (v-rel) oncogene related B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Relb | mm39_v1_chr7_-_19362328_19362398 | -0.18 | 2.9e-01 | Click! |
Activity profile of Relb motif
Sorted Z-values of Relb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_99276310 | 2.72 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr17_-_56312555 | 2.23 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr2_-_5719302 | 2.01 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr1_-_171023798 | 1.72 |
ENSMUST00000111332.2
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr17_+_56312672 | 1.68 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr6_-_124718316 | 1.62 |
ENSMUST00000004389.6
|
Grcc10
|
gene rich cluster, C10 gene |
| chr14_+_55797934 | 1.56 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr15_-_31367872 | 1.47 |
ENSMUST00000123325.9
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr15_-_31367668 | 1.45 |
ENSMUST00000110410.10
ENSMUST00000076942.5 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr15_-_98575332 | 1.44 |
ENSMUST00000120997.2
ENSMUST00000109149.9 ENSMUST00000003451.11 |
Rnd1
|
Rho family GTPase 1 |
| chr12_+_41074089 | 1.34 |
ENSMUST00000132121.8
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_41074328 | 1.28 |
ENSMUST00000134965.8
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr16_-_30086317 | 1.27 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr12_+_111132847 | 1.02 |
ENSMUST00000021706.11
|
Traf3
|
TNF receptor-associated factor 3 |
| chr12_-_54909568 | 0.99 |
ENSMUST00000078124.8
|
Cfl2
|
cofilin 2, muscle |
| chr12_+_111132779 | 0.96 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr4_-_116991150 | 0.89 |
ENSMUST00000076859.12
|
Plk3
|
polo like kinase 3 |
| chr7_+_28466160 | 0.84 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr12_+_111132908 | 0.77 |
ENSMUST00000139162.8
ENSMUST00000060274.7 |
Traf3
|
TNF receptor-associated factor 3 |
| chr17_+_37504783 | 0.77 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr18_+_53995260 | 0.74 |
ENSMUST00000237880.2
|
Csnk1g3
|
casein kinase 1, gamma 3 |
| chr12_-_55539372 | 0.70 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr5_-_121975676 | 0.69 |
ENSMUST00000197892.3
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr10_+_96452860 | 0.62 |
ENSMUST00000038377.9
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr18_+_36693024 | 0.62 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr4_-_42168603 | 0.57 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr4_-_42665763 | 0.57 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr7_-_28465870 | 0.55 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chrX_+_162691978 | 0.54 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_-_23720953 | 0.52 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr5_+_91039092 | 0.47 |
ENSMUST00000031327.9
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1 |
| chr5_-_132570710 | 0.44 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr1_+_132996237 | 0.43 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr2_+_59442378 | 0.42 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr8_+_47192767 | 0.42 |
ENSMUST00000034041.9
ENSMUST00000208507.2 ENSMUST00000207105.2 |
Irf2
|
interferon regulatory factor 2 |
| chrX_+_140258381 | 0.41 |
ENSMUST00000112931.8
ENSMUST00000112930.8 |
Col4a5
|
collagen, type IV, alpha 5 |
| chr10_-_93146937 | 0.37 |
ENSMUST00000008542.12
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr16_+_91343936 | 0.35 |
ENSMUST00000023687.9
|
Ifngr2
|
interferon gamma receptor 2 |
| chr6_+_67873135 | 0.33 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr4_-_82423985 | 0.33 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr5_-_37981919 | 0.32 |
ENSMUST00000063116.10
|
Msx1
|
msh homeobox 1 |
| chr13_+_83672389 | 0.32 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr2_+_18677195 | 0.31 |
ENSMUST00000171845.8
ENSMUST00000061158.5 |
Commd3
|
COMM domain containing 3 |
| chr2_-_160155536 | 0.30 |
ENSMUST00000109475.3
|
Gm826
|
predicted gene 826 |
| chrX_+_162692126 | 0.30 |
ENSMUST00000033734.14
ENSMUST00000112294.9 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr4_-_82423944 | 0.30 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr12_+_102915102 | 0.29 |
ENSMUST00000101099.12
|
Unc79
|
unc-79 homolog |
| chr11_+_89921121 | 0.29 |
ENSMUST00000092788.4
|
Tmem100
|
transmembrane protein 100 |
| chr4_-_91288221 | 0.29 |
ENSMUST00000102799.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr2_-_5068807 | 0.29 |
ENSMUST00000114996.8
|
Optn
|
optineurin |
| chr2_-_34848148 | 0.28 |
ENSMUST00000201690.2
ENSMUST00000172159.8 |
Traf1
|
TNF receptor-associated factor 1 |
| chr1_+_15357478 | 0.28 |
ENSMUST00000175681.3
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr16_-_17711950 | 0.28 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr10_-_127358231 | 0.27 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr10_-_127358300 | 0.25 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr10_+_102348076 | 0.24 |
ENSMUST00000219445.2
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr7_-_19363280 | 0.24 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr16_+_17712061 | 0.23 |
ENSMUST00000046937.4
|
Tssk1
|
testis-specific serine kinase 1 |
| chr10_-_93146825 | 0.23 |
ENSMUST00000151153.2
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr14_-_55828511 | 0.21 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr2_-_26917921 | 0.20 |
ENSMUST00000102890.11
ENSMUST00000153388.2 ENSMUST00000045702.6 |
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_-_5068743 | 0.20 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chrX_+_102365004 | 0.20 |
ENSMUST00000033689.3
|
Cdx4
|
caudal type homeobox 4 |
| chr4_+_126156118 | 0.20 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr17_-_71153283 | 0.19 |
ENSMUST00000156484.2
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr6_+_39848367 | 0.19 |
ENSMUST00000239008.2
|
Tmem178b
|
transmembrane protein 178B |
| chr7_-_70010341 | 0.19 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_192984278 | 0.19 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chr16_-_18052937 | 0.17 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr8_+_47192911 | 0.16 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr2_-_118377500 | 0.15 |
ENSMUST00000125860.3
|
Bmf
|
BCL2 modifying factor |
| chr1_-_135186176 | 0.15 |
ENSMUST00000185752.2
ENSMUST00000003135.14 |
Elf3
|
E74-like factor 3 |
| chr14_+_55170152 | 0.14 |
ENSMUST00000037863.6
|
Il25
|
interleukin 25 |
| chr16_+_88525719 | 0.14 |
ENSMUST00000060494.8
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr18_-_20247666 | 0.14 |
ENSMUST00000224432.2
|
Dsc1
|
desmocollin 1 |
| chr4_-_40239778 | 0.14 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr14_-_10185787 | 0.14 |
ENSMUST00000225871.2
|
Gm49355
|
predicted gene, 49355 |
| chr17_-_57456633 | 0.13 |
ENSMUST00000019633.8
|
Cd70
|
CD70 antigen |
| chr19_+_46293160 | 0.12 |
ENSMUST00000073116.13
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr19_+_45003304 | 0.08 |
ENSMUST00000039016.14
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr11_+_102775991 | 0.07 |
ENSMUST00000100369.4
|
Fam187a
|
family with sequence similarity 187, member A |
| chr8_+_106075475 | 0.07 |
ENSMUST00000073149.7
|
Slc9a5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5 |
| chr7_+_43112644 | 0.07 |
ENSMUST00000203769.2
|
Vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr9_-_77452109 | 0.06 |
ENSMUST00000183734.2
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr4_+_129713991 | 0.06 |
ENSMUST00000030578.14
|
Ptp4a2
|
protein tyrosine phosphatase 4a2 |
| chr6_-_69920632 | 0.06 |
ENSMUST00000198880.5
ENSMUST00000103371.3 |
Igkv12-38
|
immunoglobulin kappa chain variable 12-38 |
| chr10_-_18891095 | 0.05 |
ENSMUST00000019997.11
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr4_-_148584906 | 0.05 |
ENSMUST00000030840.4
|
Angptl7
|
angiopoietin-like 7 |
| chr17_+_34129221 | 0.04 |
ENSMUST00000174541.9
|
Daxx
|
Fas death domain-associated protein |
| chr4_+_151012375 | 0.04 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr2_+_119803180 | 0.03 |
ENSMUST00000066058.8
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr15_-_79048674 | 0.03 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr1_+_74830675 | 0.02 |
ENSMUST00000006718.15
|
Wnt10a
|
wingless-type MMTV integration site family, member 10A |
| chr6_+_68547717 | 0.01 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr14_+_53878403 | 0.01 |
ENSMUST00000184874.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chrX_-_133012457 | 0.00 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Relb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.6 | 1.7 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.3 | 1.6 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.2 | 2.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 2.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.2 | 1.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 0.5 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 1.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.5 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.2 | 0.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.6 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.2 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.2 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.3 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.4 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.8 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 2.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.4 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 3.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 0.5 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.1 | 0.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 2.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 2.6 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.7 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |