Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
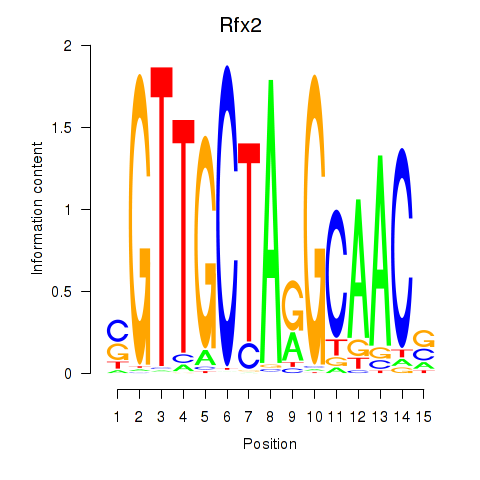
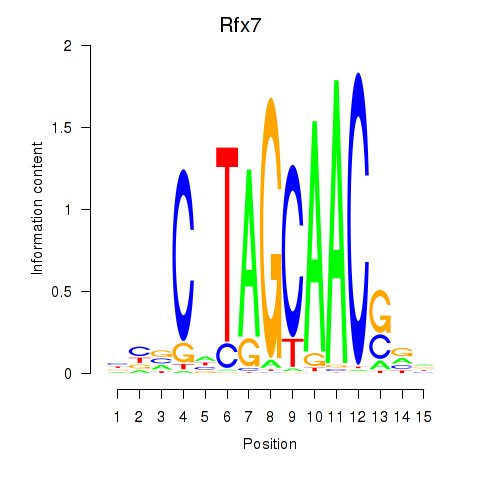
Results for Rfx2_Rfx7
Z-value: 0.90
Transcription factors associated with Rfx2_Rfx7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx2
|
ENSMUSG00000024206.16 | regulatory factor X, 2 (influences HLA class II expression) |
|
Rfx7
|
ENSMUSG00000037674.16 | regulatory factor X, 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx7 | mm39_v1_chr9_+_72439496_72439553 | -0.46 | 4.7e-03 | Click! |
| Rfx2 | mm39_v1_chr17_-_57137898_57138013 | 0.18 | 3.0e-01 | Click! |
Activity profile of Rfx2_Rfx7 motif
Sorted Z-values of Rfx2_Rfx7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_44117444 | 5.91 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr7_+_44117511 | 5.58 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr7_+_44117475 | 5.06 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr7_+_44117404 | 4.11 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr10_-_62258195 | 2.67 |
ENSMUST00000020277.9
|
Hkdc1
|
hexokinase domain containing 1 |
| chr11_+_114618209 | 1.69 |
ENSMUST00000069325.14
|
Dnai2
|
dynein axonemal intermediate chain 2 |
| chr11_-_120715351 | 1.55 |
ENSMUST00000055655.9
|
Fasn
|
fatty acid synthase |
| chr7_+_140774962 | 1.55 |
ENSMUST00000047093.11
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr2_-_152673585 | 1.49 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr7_-_4687916 | 1.42 |
ENSMUST00000206306.2
ENSMUST00000205952.2 ENSMUST00000079970.6 |
Hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr7_+_140711181 | 1.33 |
ENSMUST00000026568.10
|
Ptdss2
|
phosphatidylserine synthase 2 |
| chr8_+_112667328 | 1.31 |
ENSMUST00000034428.8
|
Gabarapl2
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 2 |
| chr1_-_131065967 | 1.20 |
ENSMUST00000189756.2
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr7_+_24246575 | 1.20 |
ENSMUST00000063249.9
|
Xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr1_-_131066004 | 1.20 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr8_-_71245036 | 1.17 |
ENSMUST00000212038.2
ENSMUST00000212551.2 ENSMUST00000211948.2 |
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr10_-_93425553 | 1.16 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr7_-_132415528 | 1.06 |
ENSMUST00000097998.9
|
Fam53b
|
family with sequence similarity 53, member B |
| chr11_-_5900019 | 1.02 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr12_-_40088024 | 0.95 |
ENSMUST00000101472.4
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr2_-_152673032 | 0.93 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr11_-_116472272 | 0.92 |
ENSMUST00000082152.5
|
Ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr15_-_81810349 | 0.91 |
ENSMUST00000023113.7
|
Polr3h
|
polymerase (RNA) III (DNA directed) polypeptide H |
| chr5_-_117527094 | 0.89 |
ENSMUST00000111953.2
ENSMUST00000086461.13 |
Rfc5
|
replication factor C (activator 1) 5 |
| chr8_+_39472981 | 0.83 |
ENSMUST00000239508.1
ENSMUST00000239509.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr11_+_102772030 | 0.78 |
ENSMUST00000021307.10
ENSMUST00000159834.2 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr7_-_44542098 | 0.78 |
ENSMUST00000003049.8
|
Med25
|
mediator complex subunit 25 |
| chr3_-_50398027 | 0.77 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr4_+_134591847 | 0.68 |
ENSMUST00000030627.8
|
Rhd
|
Rh blood group, D antigen |
| chr11_+_117006020 | 0.65 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr11_-_53371050 | 0.65 |
ENSMUST00000104955.4
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr11_-_102771751 | 0.65 |
ENSMUST00000021306.14
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr9_-_21202353 | 0.64 |
ENSMUST00000086374.8
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr9_-_21913896 | 0.64 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr4_+_6191084 | 0.63 |
ENSMUST00000029907.6
|
Ubxn2b
|
UBX domain protein 2B |
| chr9_-_21202545 | 0.63 |
ENSMUST00000215619.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr7_+_18810097 | 0.63 |
ENSMUST00000032570.14
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr2_+_162916551 | 0.62 |
ENSMUST00000142729.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr17_+_34823236 | 0.62 |
ENSMUST00000174041.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr8_+_41246310 | 0.61 |
ENSMUST00000056331.8
|
Adam20
|
a disintegrin and metallopeptidase domain 20 |
| chr2_+_32665781 | 0.60 |
ENSMUST00000066352.6
|
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr7_-_45463701 | 0.60 |
ENSMUST00000210898.2
ENSMUST00000107729.10 ENSMUST00000056820.13 |
Cyth2
|
cytohesin 2 |
| chrX_-_93410383 | 0.59 |
ENSMUST00000044989.12
|
Fam90a1b
|
family with sequence similarity 90, member A1B |
| chr9_+_72345801 | 0.59 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr5_+_30486375 | 0.59 |
ENSMUST00000101448.5
|
Drc1
|
dynein regulatory complex subunit 1 |
| chr11_-_102771806 | 0.58 |
ENSMUST00000107060.8
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr11_-_78056347 | 0.58 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr2_-_32665637 | 0.58 |
ENSMUST00000161958.2
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr10_+_79505203 | 0.57 |
ENSMUST00000020552.8
ENSMUST00000239401.2 |
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr4_+_137989526 | 0.57 |
ENSMUST00000030539.10
|
Kif17
|
kinesin family member 17 |
| chr3_-_75389047 | 0.56 |
ENSMUST00000193989.4
ENSMUST00000203169.3 |
Wdr49
|
WD repeat domain 49 |
| chr17_+_25690538 | 0.56 |
ENSMUST00000234449.2
ENSMUST00000025002.3 ENSMUST00000235033.2 |
Tekt4
|
tektin 4 |
| chr11_+_117005958 | 0.56 |
ENSMUST00000021177.15
|
Sec14l1
|
SEC14-like lipid binding 1 |
| chr9_-_21202693 | 0.55 |
ENSMUST00000213407.2
|
Cdkn2d
|
cyclin dependent kinase inhibitor 2D |
| chr2_-_93292800 | 0.54 |
ENSMUST00000028644.11
|
Cd82
|
CD82 antigen |
| chr8_-_106434565 | 0.54 |
ENSMUST00000013299.11
|
Enkd1
|
enkurin domain containing 1 |
| chr19_+_9979033 | 0.54 |
ENSMUST00000121418.8
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr4_+_137989783 | 0.53 |
ENSMUST00000105821.3
|
Kif17
|
kinesin family member 17 |
| chr9_-_21913833 | 0.53 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr2_-_32665596 | 0.53 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr19_-_10460238 | 0.53 |
ENSMUST00000235392.2
ENSMUST00000237522.2 ENSMUST00000038842.5 |
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr5_-_21850579 | 0.51 |
ENSMUST00000051358.11
|
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr19_+_47926086 | 0.51 |
ENSMUST00000238163.2
ENSMUST00000066308.9 |
Cfap58
|
cilia and flagella associated protein 58 |
| chr8_+_41205245 | 0.51 |
ENSMUST00000096663.5
|
Adam25
|
a disintegrin and metallopeptidase domain 25 (testase 2) |
| chr5_-_21850539 | 0.51 |
ENSMUST00000115234.2
|
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr13_-_55684317 | 0.50 |
ENSMUST00000021956.9
ENSMUST00000224765.2 |
Ddx41
|
DEAD box helicase 41 |
| chr7_+_100145192 | 0.49 |
ENSMUST00000133044.3
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr8_-_123170102 | 0.49 |
ENSMUST00000006692.6
|
Mvd
|
mevalonate (diphospho) decarboxylase |
| chr7_-_125090540 | 0.48 |
ENSMUST00000138616.3
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr1_+_46105898 | 0.48 |
ENSMUST00000069293.10
|
Dnah7b
|
dynein, axonemal, heavy chain 7B |
| chr1_-_133728779 | 0.47 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr7_+_44545501 | 0.47 |
ENSMUST00000071207.14
ENSMUST00000209132.2 ENSMUST00000207069.2 ENSMUST00000209039.2 ENSMUST00000207939.3 ENSMUST00000207485.2 ENSMUST00000208179.2 |
Fuz
|
fuzzy planar cell polarity protein |
| chr2_+_70339157 | 0.46 |
ENSMUST00000100041.9
|
Erich2
|
glutamate rich 2 |
| chr7_-_125090757 | 0.46 |
ENSMUST00000033006.14
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr2_+_30331839 | 0.46 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr2_+_70339175 | 0.46 |
ENSMUST00000134607.8
|
Erich2
|
glutamate rich 2 |
| chr14_+_55855484 | 0.45 |
ENSMUST00000002395.8
|
Rec8
|
REC8 meiotic recombination protein |
| chr7_+_4925781 | 0.45 |
ENSMUST00000207527.2
ENSMUST00000207687.2 ENSMUST00000208754.2 |
Nat14
|
N-acetyltransferase 14 |
| chr11_-_66059330 | 0.45 |
ENSMUST00000080665.10
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr11_-_66059270 | 0.45 |
ENSMUST00000108691.2
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr17_+_69144053 | 0.44 |
ENSMUST00000178545.3
|
Tmem200c
|
transmembrane protein 200C |
| chr11_+_101358990 | 0.44 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr2_-_151822114 | 0.44 |
ENSMUST00000062047.6
|
Fam110a
|
family with sequence similarity 110, member A |
| chr14_-_56339915 | 0.44 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr7_+_26932425 | 0.44 |
ENSMUST00000003860.13
ENSMUST00000108378.10 |
Coq8b
|
coenzyme Q8B |
| chr11_+_100436433 | 0.43 |
ENSMUST00000092684.12
ENSMUST00000006976.8 |
Odad4
|
outer dynein arm complex subunit 4 |
| chr1_+_46106006 | 0.43 |
ENSMUST00000238212.2
|
Dnah7b
|
dynein, axonemal, heavy chain 7B |
| chr17_+_71980249 | 0.43 |
ENSMUST00000097284.10
|
Togaram2
|
TOG array regulator of axonemal microtubules 2 |
| chr14_+_46997984 | 0.42 |
ENSMUST00000067426.6
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr11_+_78068931 | 0.42 |
ENSMUST00000147819.8
|
Tlcd1
|
TLC domain containing 1 |
| chr7_+_126575510 | 0.42 |
ENSMUST00000206780.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr5_-_108922819 | 0.42 |
ENSMUST00000200159.2
ENSMUST00000212212.2 |
Rnf212
|
ring finger protein 212 |
| chr4_-_117109074 | 0.42 |
ENSMUST00000165128.9
|
Armh1
|
armadillo-like helical domain containing 1 |
| chr6_-_87699767 | 0.42 |
ENSMUST00000238521.2
|
1810020O05Rik
|
Riken cDNA 1810020O05 gene |
| chr7_-_55669702 | 0.42 |
ENSMUST00000052204.6
|
Nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) |
| chr8_-_85751897 | 0.42 |
ENSMUST00000064314.10
|
Get3
|
guided entry of tail-anchored proteins factor 3, ATPase |
| chr15_-_54953819 | 0.41 |
ENSMUST00000110231.2
ENSMUST00000023059.13 |
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr12_+_111132779 | 0.41 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr3_+_82265351 | 0.40 |
ENSMUST00000193559.6
ENSMUST00000192595.6 ENSMUST00000091014.10 |
Map9
|
microtubule-associated protein 9 |
| chr9_+_22322802 | 0.40 |
ENSMUST00000058868.9
|
9530077C05Rik
|
RIKEN cDNA 9530077C05 gene |
| chr3_+_146110709 | 0.39 |
ENSMUST00000129978.2
|
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chrX_+_71962157 | 0.39 |
ENSMUST00000033715.5
|
Nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr11_+_68477812 | 0.39 |
ENSMUST00000154294.8
ENSMUST00000063006.12 |
Ccdc42
|
coiled-coil domain containing 42 |
| chr10_+_57521930 | 0.39 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr11_+_97697128 | 0.38 |
ENSMUST00000138919.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr17_-_11059172 | 0.38 |
ENSMUST00000041463.7
|
Pacrg
|
PARK2 co-regulated |
| chr9_+_72345267 | 0.38 |
ENSMUST00000183809.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr8_-_79975199 | 0.37 |
ENSMUST00000034109.6
|
1700011L22Rik
|
RIKEN cDNA 1700011L22 gene |
| chr8_-_55177510 | 0.37 |
ENSMUST00000175915.8
|
Wdr17
|
WD repeat domain 17 |
| chr17_-_46558894 | 0.37 |
ENSMUST00000142706.9
ENSMUST00000173349.8 ENSMUST00000087026.13 |
Polr1c
|
polymerase (RNA) I polypeptide C |
| chr7_-_28465870 | 0.37 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr11_-_117673008 | 0.37 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr1_-_16163506 | 0.37 |
ENSMUST00000145070.8
ENSMUST00000151004.2 |
4930444P10Rik
|
RIKEN cDNA 4930444P10 gene |
| chr12_-_85335193 | 0.37 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr1_-_119764729 | 0.37 |
ENSMUST00000163621.2
ENSMUST00000168303.8 |
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr16_+_36832119 | 0.37 |
ENSMUST00000071452.12
ENSMUST00000054034.7 |
Polq
|
polymerase (DNA directed), theta |
| chr3_+_156267429 | 0.37 |
ENSMUST00000074015.11
|
Negr1
|
neuronal growth regulator 1 |
| chr5_-_149559667 | 0.36 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr9_+_49014020 | 0.36 |
ENSMUST00000070390.12
ENSMUST00000167095.8 |
Tmprss5
|
transmembrane protease, serine 5 (spinesin) |
| chr11_+_115802828 | 0.36 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr5_-_116427003 | 0.36 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr8_+_72993862 | 0.36 |
ENSMUST00000003117.15
ENSMUST00000212841.2 |
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr7_+_18810167 | 0.35 |
ENSMUST00000108479.2
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr4_+_100336003 | 0.35 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr5_+_30623379 | 0.35 |
ENSMUST00000127815.2
ENSMUST00000031078.10 ENSMUST00000114743.2 |
Fam166c
|
family with sequence similarity 166, member C |
| chr9_-_21061196 | 0.35 |
ENSMUST00000215296.2
ENSMUST00000019615.11 |
Cdc37
|
cell division cycle 37 |
| chr14_-_64225223 | 0.35 |
ENSMUST00000022532.6
|
4930578I06Rik
|
RIKEN cDNA 4930578I06 gene |
| chr8_-_55177352 | 0.35 |
ENSMUST00000129132.3
ENSMUST00000150488.8 ENSMUST00000127511.9 |
Wdr17
|
WD repeat domain 17 |
| chr11_-_69470139 | 0.34 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr11_+_62441992 | 0.34 |
ENSMUST00000019649.4
|
Ubb
|
ubiquitin B |
| chr7_-_79882313 | 0.34 |
ENSMUST00000206084.2
ENSMUST00000205996.2 ENSMUST00000071457.12 |
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr12_+_111132847 | 0.34 |
ENSMUST00000021706.11
|
Traf3
|
TNF receptor-associated factor 3 |
| chr11_-_30421792 | 0.34 |
ENSMUST00000041763.14
|
4930505A04Rik
|
RIKEN cDNA 4930505A04 gene |
| chr3_+_82265474 | 0.34 |
ENSMUST00000195471.6
ENSMUST00000195640.2 |
Map9
|
microtubule-associated protein 9 |
| chr3_+_146110387 | 0.33 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr6_-_38852857 | 0.33 |
ENSMUST00000162359.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr1_-_53745920 | 0.33 |
ENSMUST00000094964.7
|
Dnah7a
|
dynein, axonemal, heavy chain 7A |
| chr9_+_119766672 | 0.32 |
ENSMUST00000035100.6
|
Ttc21a
|
tetratricopeptide repeat domain 21A |
| chr6_-_8259098 | 0.32 |
ENSMUST00000012627.5
|
Rpa3
|
replication protein A3 |
| chr15_+_78798116 | 0.32 |
ENSMUST00000089378.5
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr6_+_128352419 | 0.32 |
ENSMUST00000130785.4
ENSMUST00000100926.4 ENSMUST00000204223.2 |
Tex52
Gm44596
|
testis expressed 52 predicted gene 44596 |
| chr11_+_59839032 | 0.32 |
ENSMUST00000081980.7
|
Med9
|
mediator complex subunit 9 |
| chr6_-_38852899 | 0.32 |
ENSMUST00000160360.2
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr14_+_46998004 | 0.32 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr2_-_35226981 | 0.32 |
ENSMUST00000028241.7
|
Stom
|
stomatin |
| chr12_+_87247297 | 0.32 |
ENSMUST00000182869.2
|
Samd15
|
sterile alpha motif domain containing 15 |
| chr5_-_149559792 | 0.32 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_+_75655504 | 0.32 |
ENSMUST00000189155.4
|
Gm29133
|
predicted gene 29133 |
| chr5_+_120727068 | 0.31 |
ENSMUST00000069259.9
ENSMUST00000094391.6 |
Iqcd
|
IQ motif containing D |
| chr1_-_119765068 | 0.31 |
ENSMUST00000163435.8
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr12_+_113149487 | 0.31 |
ENSMUST00000058491.8
|
Tmem121
|
transmembrane protein 121 |
| chr10_+_57521958 | 0.31 |
ENSMUST00000177473.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr17_-_34822649 | 0.31 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr12_+_33444853 | 0.31 |
ENSMUST00000020878.8
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr6_-_113577606 | 0.31 |
ENSMUST00000035870.5
|
Fancd2os
|
Fancd2 opposite strand |
| chr1_+_60448931 | 0.30 |
ENSMUST00000189082.7
ENSMUST00000187709.7 |
Abi2
|
abl interactor 2 |
| chr5_+_27466914 | 0.30 |
ENSMUST00000101471.4
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr7_-_143153785 | 0.30 |
ENSMUST00000105909.4
ENSMUST00000010899.14 |
Cars
|
cysteinyl-tRNA synthetase |
| chr7_+_4785936 | 0.30 |
ENSMUST00000013235.6
|
Tmem190
|
transmembrane protein 190 |
| chr11_+_11414256 | 0.29 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr5_+_123627365 | 0.29 |
ENSMUST00000196809.5
ENSMUST00000094327.10 ENSMUST00000121444.2 |
Lrrc43
|
leucine rich repeat containing 43 |
| chr10_-_22607136 | 0.29 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr4_+_118266526 | 0.29 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr2_-_164876690 | 0.29 |
ENSMUST00000122070.2
ENSMUST00000121377.8 ENSMUST00000153905.2 ENSMUST00000040381.15 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr18_+_57666852 | 0.29 |
ENSMUST00000079738.10
ENSMUST00000135806.8 ENSMUST00000127130.9 |
Ccdc192
|
coiled-coil domain containing 192 |
| chr2_-_181340226 | 0.29 |
ENSMUST00000052416.4
|
Lkaaear1
|
LKAAEAR motif containing 1 (IKAAEAR murine motif) |
| chr11_+_117672902 | 0.29 |
ENSMUST00000127080.9
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr9_-_103569984 | 0.29 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr2_-_69619864 | 0.29 |
ENSMUST00000094942.4
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr7_-_28297565 | 0.28 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr2_-_152239966 | 0.28 |
ENSMUST00000063332.9
ENSMUST00000182625.2 |
Sox12
|
SRY (sex determining region Y)-box 12 |
| chr5_+_124802149 | 0.28 |
ENSMUST00000141137.7
ENSMUST00000058440.12 |
Dnah10
|
dynein, axonemal, heavy chain 10 |
| chr6_+_85564566 | 0.28 |
ENSMUST00000213058.2
|
Alms1
|
ALMS1, centrosome and basal body associated |
| chr10_-_74868360 | 0.28 |
ENSMUST00000159994.2
ENSMUST00000179546.8 ENSMUST00000160450.8 ENSMUST00000160072.8 ENSMUST00000009214.10 ENSMUST00000166088.8 |
Rsph14
|
radial spoke head homolog 14 (Chlamydomonas) |
| chr8_+_27937691 | 0.28 |
ENSMUST00000081321.5
|
Poteg
|
POTE ankyrin domain family, member G |
| chr5_+_135807334 | 0.27 |
ENSMUST00000019323.11
|
Mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr2_-_71377088 | 0.27 |
ENSMUST00000024159.8
|
Dlx2
|
distal-less homeobox 2 |
| chr5_-_149559636 | 0.27 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr15_+_85016261 | 0.27 |
ENSMUST00000023067.4
|
Ribc2
|
RIB43A domain with coiled-coils 2 |
| chr7_+_90075762 | 0.27 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr11_-_119190896 | 0.27 |
ENSMUST00000026667.15
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr7_-_79882501 | 0.27 |
ENSMUST00000065163.15
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr8_+_106434901 | 0.27 |
ENSMUST00000013302.7
ENSMUST00000211852.2 |
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr19_+_34078333 | 0.26 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr6_-_24515036 | 0.26 |
ENSMUST00000052277.5
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chr3_+_156267587 | 0.26 |
ENSMUST00000041425.12
ENSMUST00000106065.2 |
Negr1
|
neuronal growth regulator 1 |
| chr1_+_191553556 | 0.26 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr5_+_77163869 | 0.26 |
ENSMUST00000031161.11
ENSMUST00000117880.8 |
Thegl
|
theg spermatid protein like |
| chr14_-_29443792 | 0.26 |
ENSMUST00000022567.9
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr8_+_123844090 | 0.25 |
ENSMUST00000037900.9
|
Cpne7
|
copine VII |
| chr15_-_89263466 | 0.25 |
ENSMUST00000228111.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr7_-_127187767 | 0.25 |
ENSMUST00000072155.5
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr7_-_44541787 | 0.25 |
ENSMUST00000208551.2
ENSMUST00000208253.2 ENSMUST00000207654.2 ENSMUST00000207278.2 |
Med25
|
mediator complex subunit 25 |
| chr7_+_3632982 | 0.25 |
ENSMUST00000179769.8
ENSMUST00000008517.13 |
Prpf31
|
pre-mRNA processing factor 31 |
| chr6_+_83055321 | 0.25 |
ENSMUST00000165164.9
ENSMUST00000092614.9 |
Pcgf1
|
polycomb group ring finger 1 |
| chr16_-_3725515 | 0.25 |
ENSMUST00000177221.2
ENSMUST00000177323.8 |
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr5_+_98477157 | 0.25 |
ENSMUST00000080333.8
|
Cfap299
|
cilia and flagella associated protein 299 |
| chr12_+_113120023 | 0.25 |
ENSMUST00000049271.13
|
Tedc1
|
tubulin epsilon and delta complex 1 |
| chr4_+_118266582 | 0.25 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr6_-_73198608 | 0.25 |
ENSMUST00000064948.13
ENSMUST00000114040.8 |
Dnah6
|
dynein, axonemal, heavy chain 6 |
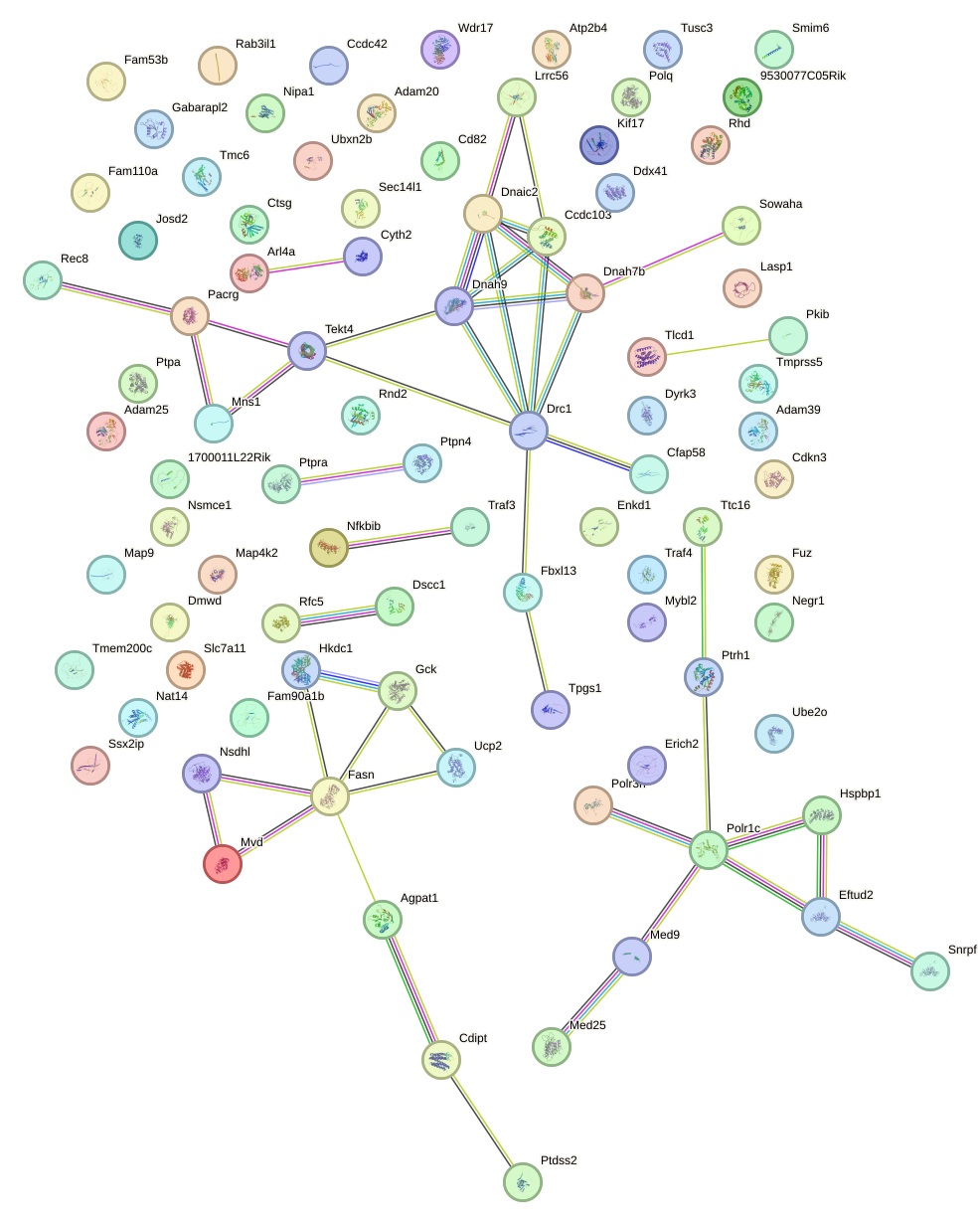
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx2_Rfx7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.5 | 2.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 1.0 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.2 | 1.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.2 | 1.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.2 | 1.6 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 0.8 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.2 | 1.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.5 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.2 | 0.5 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 1.1 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.2 | 0.5 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.2 | 2.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.7 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 21.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.8 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.5 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.3 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.1 | 0.6 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.1 | 0.3 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.1 | 2.4 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 0.3 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.3 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.4 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 1.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.2 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.3 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.3 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 2.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.4 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.2 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.9 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.4 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 1.2 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0006348 | DNA replication-dependent nucleosome assembly(GO:0006335) chromatin silencing at telomere(GO:0006348) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) termination of signal transduction(GO:0023021) |
| 0.0 | 0.2 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.8 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.1 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 2.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.2 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 2.0 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.3 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 1.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.4 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0031177 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-acetyltransferase activity(GO:0016418) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) phosphopantetheine binding(GO:0031177) |
| 0.4 | 3.7 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.2 | 2.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 20.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 0.7 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.8 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 1.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.7 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 2.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.3 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.2 | GO:0008456 | alpha-N-acetylgalactosaminidase activity(GO:0008456) |
| 0.1 | 0.3 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.1 | 0.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 2.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 1.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |