Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
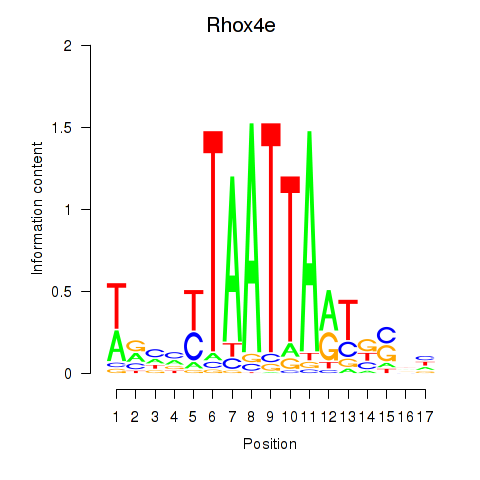
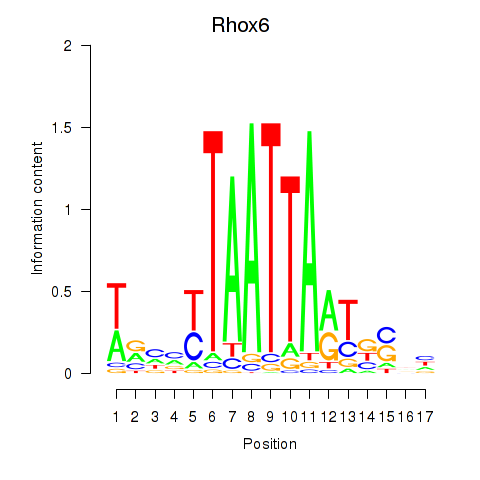
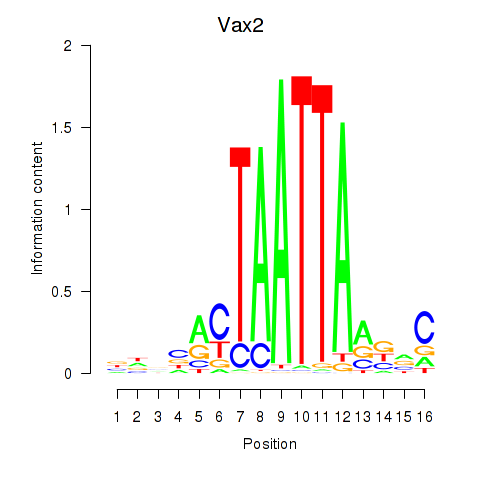
Results for Rhox4e_Rhox6_Vax2
Z-value: 0.65
Transcription factors associated with Rhox4e_Rhox6_Vax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rhox4e
|
ENSMUSG00000071770.6 | reproductive homeobox 4E |
|
Rhox6
|
ENSMUSG00000006200.4 | reproductive homeobox 6 |
|
Vax2
|
ENSMUSG00000034777.3 | ventral anterior homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rhox4e | mm39_v1_chrX_+_36739065_36739065 | -0.15 | 3.9e-01 | Click! |
| Vax2 | mm39_v1_chr6_+_83688213_83688270 | 0.14 | 4.0e-01 | Click! |
Activity profile of Rhox4e_Rhox6_Vax2 motif
Sorted Z-values of Rhox4e_Rhox6_Vax2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_127734384 | 5.05 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr7_+_51528788 | 5.01 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr6_+_121815473 | 4.26 |
ENSMUST00000032228.9
|
Mug1
|
murinoglobulin 1 |
| chr19_-_39637489 | 4.09 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr3_-_67422821 | 3.76 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr16_+_22737128 | 3.64 |
ENSMUST00000170805.9
|
Fetub
|
fetuin beta |
| chr16_+_22737227 | 3.54 |
ENSMUST00000231880.2
|
Fetub
|
fetuin beta |
| chr16_+_22737050 | 3.50 |
ENSMUST00000231768.2
|
Fetub
|
fetuin beta |
| chr8_+_46111703 | 3.48 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr4_+_43493344 | 3.41 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr6_+_121983720 | 3.36 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr15_+_59186876 | 2.74 |
ENSMUST00000022977.14
ENSMUST00000100640.5 |
Sqle
|
squalene epoxidase |
| chr11_-_113600838 | 2.41 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr2_+_69727563 | 2.20 |
ENSMUST00000055758.16
ENSMUST00000112251.9 |
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr5_-_87240405 | 2.06 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr2_+_172994841 | 1.80 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr19_-_39801188 | 1.79 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr2_+_69727599 | 1.66 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr13_+_24023428 | 1.65 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr5_+_90708962 | 1.57 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr12_-_79237722 | 1.52 |
ENSMUST00000085254.7
|
Rdh11
|
retinol dehydrogenase 11 |
| chr2_-_154916367 | 1.40 |
ENSMUST00000137242.2
ENSMUST00000054607.16 |
Ahcy
|
S-adenosylhomocysteine hydrolase |
| chrX_+_102400061 | 1.36 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr13_+_24023386 | 1.24 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr14_-_45626237 | 1.23 |
ENSMUST00000227865.2
ENSMUST00000226856.2 ENSMUST00000226276.2 ENSMUST00000046191.9 |
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr7_-_12829100 | 1.22 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chrM_+_8603 | 1.20 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr8_+_45960804 | 1.20 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr13_-_53627110 | 1.20 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr5_-_87402659 | 1.13 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr18_-_10706701 | 1.09 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr11_+_58062467 | 1.07 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr7_-_126275529 | 1.04 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chrM_+_10167 | 1.04 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr4_+_100336003 | 1.01 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr8_+_45960931 | 1.00 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_125357756 | 1.00 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr10_-_81243475 | 1.00 |
ENSMUST00000140916.8
|
Nfic
|
nuclear factor I/C |
| chr9_+_21634779 | 0.96 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr4_-_14621805 | 0.95 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr18_+_60515755 | 0.95 |
ENSMUST00000237185.2
|
Iigp1
|
interferon inducible GTPase 1 |
| chr9_-_71070506 | 0.94 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr1_+_87983099 | 0.93 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr10_-_126956991 | 0.90 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr18_+_60345152 | 0.90 |
ENSMUST00000031549.6
|
Gm4951
|
predicted gene 4951 |
| chr18_+_12874390 | 0.90 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr1_-_72323464 | 0.88 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr1_-_72323407 | 0.87 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr2_+_163348728 | 0.87 |
ENSMUST00000143911.8
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr4_-_14621669 | 0.86 |
ENSMUST00000143105.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr4_-_129121676 | 0.81 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr2_-_132089667 | 0.81 |
ENSMUST00000110163.8
ENSMUST00000180286.2 ENSMUST00000028816.9 |
Tmem230
|
transmembrane protein 230 |
| chr10_+_127226180 | 0.81 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chrM_+_7758 | 0.80 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr8_+_46111778 | 0.80 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr5_+_104350475 | 0.78 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr1_+_87983189 | 0.77 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr9_+_80072361 | 0.76 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr17_-_59320257 | 0.76 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr17_+_79919267 | 0.74 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chrM_+_9870 | 0.73 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr15_-_74508197 | 0.73 |
ENSMUST00000023271.8
|
Mroh4
|
maestro heat-like repeat family member 4 |
| chr9_-_50472605 | 0.72 |
ENSMUST00000034568.7
|
Tex12
|
testis expressed 12 |
| chr14_-_45626198 | 0.72 |
ENSMUST00000226590.2
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr7_-_28855143 | 0.72 |
ENSMUST00000153251.8
|
Fam98c
|
family with sequence similarity 98, member C |
| chr4_-_133066594 | 0.72 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chrM_+_7779 | 0.71 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chrM_+_2743 | 0.70 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr9_-_50472620 | 0.69 |
ENSMUST00000217236.2
|
Tex12
|
testis expressed 12 |
| chr9_-_65792306 | 0.64 |
ENSMUST00000122410.8
ENSMUST00000117083.2 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr18_+_38551960 | 0.63 |
ENSMUST00000236085.2
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr16_+_18655318 | 0.62 |
ENSMUST00000055413.13
ENSMUST00000123146.8 ENSMUST00000191388.2 |
2510002D24Rik
|
RIKEN cDNA 2510002D24 gene |
| chr13_-_103042294 | 0.60 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_-_39875192 | 0.58 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr14_-_68771138 | 0.58 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr19_+_60800012 | 0.58 |
ENSMUST00000128357.8
ENSMUST00000119633.8 ENSMUST00000025957.9 |
Dennd10
|
DENN domain containing 10 |
| chr13_-_17869314 | 0.57 |
ENSMUST00000221598.2
ENSMUST00000068545.6 ENSMUST00000220514.2 |
Sugct
|
succinyl-CoA glutarate-CoA transferase |
| chr18_+_12874368 | 0.56 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chrM_+_14138 | 0.55 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr18_+_38552011 | 0.55 |
ENSMUST00000025293.5
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr19_-_44095840 | 0.55 |
ENSMUST00000119591.2
ENSMUST00000026217.11 |
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chrX_+_106299484 | 0.54 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chrM_+_7006 | 0.53 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr6_-_115569504 | 0.52 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr7_-_28855020 | 0.52 |
ENSMUST00000123416.2
|
Fam98c
|
family with sequence similarity 98, member C |
| chr15_-_66985760 | 0.50 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr7_+_28869770 | 0.50 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr4_-_14621497 | 0.50 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr10_+_29074950 | 0.49 |
ENSMUST00000217011.2
|
Gm49353
|
predicted gene, 49353 |
| chr5_-_3697806 | 0.49 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr3_+_121746862 | 0.48 |
ENSMUST00000037958.14
ENSMUST00000196904.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr5_-_118382926 | 0.47 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr10_+_23770586 | 0.47 |
ENSMUST00000041416.8
|
Vnn1
|
vanin 1 |
| chr13_-_103042554 | 0.47 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_24423715 | 0.46 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr6_-_41752111 | 0.46 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr12_-_85871281 | 0.45 |
ENSMUST00000021676.12
ENSMUST00000142331.2 |
Erg28
|
ergosterol biosynthesis 28 |
| chr7_+_28869629 | 0.45 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin |
| chr9_+_108216466 | 0.45 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr8_+_45960855 | 0.45 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr12_+_80690985 | 0.44 |
ENSMUST00000219405.2
ENSMUST00000085245.7 |
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr6_+_145879839 | 0.43 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr4_+_15881256 | 0.43 |
ENSMUST00000029876.2
|
Calb1
|
calbindin 1 |
| chr12_-_84664001 | 0.42 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr16_-_29363671 | 0.42 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr2_-_69542805 | 0.42 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr1_+_163757334 | 0.41 |
ENSMUST00000027876.11
ENSMUST00000170359.8 |
Scyl3
|
SCY1-like 3 (S. cerevisiae) |
| chr12_+_55350023 | 0.41 |
ENSMUST00000184766.8
ENSMUST00000183475.8 ENSMUST00000183654.2 |
Prorp
|
protein only RNase P catalytic subunit |
| chr1_+_88234454 | 0.40 |
ENSMUST00000040210.14
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr7_-_46316767 | 0.40 |
ENSMUST00000168335.3
ENSMUST00000107669.9 |
Tph1
|
tryptophan hydroxylase 1 |
| chr7_+_130375799 | 0.40 |
ENSMUST00000048453.7
ENSMUST00000208593.2 |
Btbd16
|
BTB (POZ) domain containing 16 |
| chr19_+_3373285 | 0.39 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr9_+_108216433 | 0.39 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr11_-_99241924 | 0.37 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
| chr10_-_44024843 | 0.37 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr9_-_55419442 | 0.37 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr13_-_81859056 | 0.36 |
ENSMUST00000161920.2
ENSMUST00000048993.12 |
Polr3g
|
polymerase (RNA) III (DNA directed) polypeptide G |
| chr12_+_55349422 | 0.36 |
ENSMUST00000021411.15
|
Prorp
|
protein only RNase P catalytic subunit |
| chr19_+_25384024 | 0.35 |
ENSMUST00000146647.3
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chrM_-_14061 | 0.35 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr11_+_50917831 | 0.35 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr15_+_98350469 | 0.33 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281 |
| chr2_+_177760959 | 0.31 |
ENSMUST00000108916.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr13_+_90237824 | 0.31 |
ENSMUST00000012566.9
|
Tmem167
|
transmembrane protein 167 |
| chr4_+_108576846 | 0.31 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr9_+_72866067 | 0.30 |
ENSMUST00000098567.9
ENSMUST00000034734.9 |
Dnaaf4
|
dynein axonemal assembly factor 4 |
| chr1_-_63215952 | 0.30 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr3_-_54962899 | 0.30 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr7_+_30157704 | 0.30 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr2_+_127696548 | 0.30 |
ENSMUST00000028859.8
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr2_+_121787131 | 0.29 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr1_+_173964312 | 0.29 |
ENSMUST00000053941.4
|
Olfr424
|
olfactory receptor 424 |
| chr19_-_7943365 | 0.29 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chrX_-_158921370 | 0.29 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr4_+_122910382 | 0.29 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr5_-_90788323 | 0.28 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr8_-_62355690 | 0.28 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr5_-_84565218 | 0.28 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr9_+_108216233 | 0.28 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr5_+_29400981 | 0.28 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr12_+_116239006 | 0.27 |
ENSMUST00000090195.5
|
Gm11027
|
predicted gene 11027 |
| chr1_+_135768409 | 0.27 |
ENSMUST00000189826.7
|
Tnnt2
|
troponin T2, cardiac |
| chr7_+_114344920 | 0.27 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chrX_+_149372903 | 0.26 |
ENSMUST00000080884.11
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr2_+_121786892 | 0.26 |
ENSMUST00000110578.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr4_-_150998857 | 0.26 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr5_-_66211842 | 0.25 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr13_-_43634695 | 0.25 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr11_-_117764258 | 0.25 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr16_+_56024676 | 0.25 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr14_-_36820304 | 0.25 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr17_+_14087827 | 0.24 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr14_-_17305238 | 0.23 |
ENSMUST00000179659.2
|
Gm3317
|
predicted gene 3317 |
| chr1_+_54289833 | 0.23 |
ENSMUST00000027128.11
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr14_-_17955015 | 0.23 |
ENSMUST00000112776.4
|
Gm3173
|
predicted gene 3173 |
| chr16_+_22965286 | 0.23 |
ENSMUST00000023593.6
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr3_+_103482547 | 0.23 |
ENSMUST00000121834.8
|
Syt6
|
synaptotagmin VI |
| chr5_-_143279378 | 0.23 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr2_+_144112798 | 0.23 |
ENSMUST00000028910.9
ENSMUST00000110027.2 |
Mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr8_+_114362419 | 0.22 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr14_-_19619621 | 0.22 |
ENSMUST00000112595.3
|
Gm2237
|
predicted gene 2237 |
| chr8_+_114362181 | 0.21 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr1_-_63215812 | 0.21 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr11_+_115197980 | 0.21 |
ENSMUST00000055490.9
|
Otop2
|
otopetrin 2 |
| chr1_-_144052997 | 0.21 |
ENSMUST00000111941.2
ENSMUST00000052375.8 |
Rgs13
|
regulator of G-protein signaling 13 |
| chr13_+_67981349 | 0.20 |
ENSMUST00000222626.2
ENSMUST00000060609.8 |
Gm10037
|
predicted gene 10037 |
| chr18_+_24737009 | 0.20 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr5_-_137015683 | 0.20 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr8_+_65399831 | 0.20 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr11_+_69729340 | 0.19 |
ENSMUST00000133967.8
ENSMUST00000094065.5 |
Tmem256
|
transmembrane protein 256 |
| chr11_+_116734104 | 0.19 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr1_-_135211813 | 0.19 |
ENSMUST00000077340.14
ENSMUST00000074357.8 |
Rnpep
|
arginyl aminopeptidase (aminopeptidase B) |
| chr6_-_68968278 | 0.19 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr12_+_100076407 | 0.19 |
ENSMUST00000021595.10
|
Psmc1
|
protease (prosome, macropain) 26S subunit, ATPase 1 |
| chr12_+_80691275 | 0.19 |
ENSMUST00000217889.2
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chrX_-_142716085 | 0.18 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr16_-_19341016 | 0.18 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr8_-_41494890 | 0.18 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_+_65676111 | 0.18 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr17_-_45970238 | 0.17 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr10_-_83484467 | 0.16 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr12_+_78240999 | 0.16 |
ENSMUST00000211288.3
|
Gm6657
|
predicted gene 6657 |
| chr11_-_99907030 | 0.16 |
ENSMUST00000018399.3
|
Krt33a
|
keratin 33A |
| chr8_+_46463633 | 0.16 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr16_+_22965330 | 0.16 |
ENSMUST00000171309.2
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr6_-_128599779 | 0.16 |
ENSMUST00000203275.3
|
Klrb1a
|
killer cell lectin-like receptor subfamily B member 1A |
| chr7_+_126549692 | 0.16 |
ENSMUST00000106335.8
ENSMUST00000146017.3 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr3_+_55369149 | 0.15 |
ENSMUST00000199585.5
ENSMUST00000070418.9 |
Dclk1
|
doublecortin-like kinase 1 |
| chr1_-_131441962 | 0.15 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr5_-_90788460 | 0.15 |
ENSMUST00000202704.4
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_+_41520737 | 0.15 |
ENSMUST00000103298.2
|
Trbj2-7
|
T cell receptor beta joining 2-7 |
| chrX_+_159551009 | 0.14 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr5_-_146521629 | 0.14 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr15_+_102927366 | 0.14 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr8_+_20478993 | 0.14 |
ENSMUST00000211848.2
|
Gm31371
|
predicted gene, 31371 |
| chr10_-_129965752 | 0.14 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr6_-_90055488 | 0.14 |
ENSMUST00000203791.3
ENSMUST00000226368.2 |
Vmn1r49
|
vomeronasal 1, receptor 49 |
| chr1_+_107350411 | 0.14 |
ENSMUST00000086690.6
|
Serpinb7
|
serine (or cysteine) peptidase inhibitor, clade B, member 7 |
| chr1_+_173924457 | 0.13 |
ENSMUST00000213832.2
|
Olfr427
|
olfactory receptor 427 |
| chr14_+_53315909 | 0.12 |
ENSMUST00000103608.4
|
Trav14d-3-dv8
|
T cell receptor alpha variable 14D-3-DV8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox4e_Rhox6_Vax2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.6 | 1.8 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.4 | 1.8 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.4 | 1.8 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.4 | 1.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.4 | 2.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.3 | 1.4 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.3 | 0.9 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.3 | 1.1 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.3 | 2.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 1.0 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 1.4 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 0.9 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.2 | 2.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 3.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 10.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 1.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 0.8 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 6.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 0.5 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 1.7 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.4 | GO:0072249 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 0.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.8 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.4 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 6.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.6 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:0036471 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.6 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 5.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.3 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.4 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 1.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.8 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 2.7 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 1.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.9 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.7 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 1.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.5 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.4 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 1.1 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 4.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 1.0 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0071803 | keratinocyte development(GO:0003334) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 3.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 1.0 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 1.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 1.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.4 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 2.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.9 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.4 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 1.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.3 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 1.0 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 4.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 5.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 4.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.5 | 10.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 1.8 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 0.9 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.3 | 2.9 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.3 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 5.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 1.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 0.9 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.2 | 1.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 6.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 2.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.4 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 4.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.6 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.3 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.3 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.3 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 6.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 2.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 1.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 2.0 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 4.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 4.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0009055 | electron carrier activity(GO:0009055) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |