Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Rorc_Nr1d1
Z-value: 0.53
Transcription factors associated with Rorc_Nr1d1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
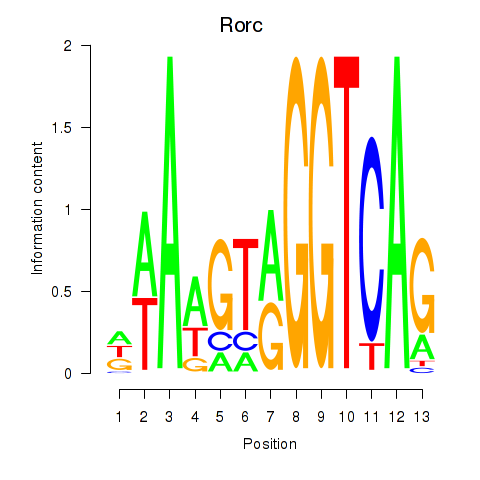
Rorc
|
ENSMUSG00000028150.15 | RAR-related orphan receptor gamma |
|
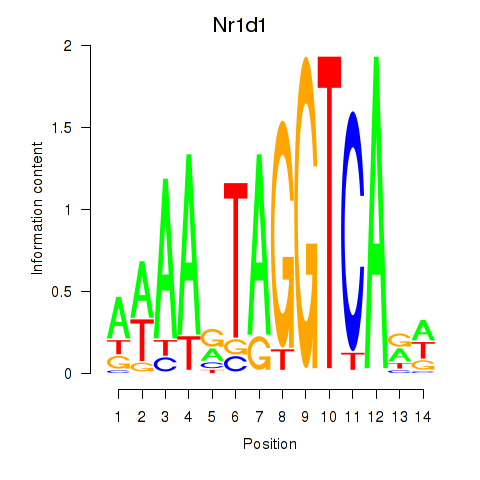
Nr1d1
|
ENSMUSG00000020889.12 | nuclear receptor subfamily 1, group D, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rorc | mm39_v1_chr3_+_94284739_94284758 | 0.11 | 5.4e-01 | Click! |
| Nr1d1 | mm39_v1_chr11_-_98666159_98666183 | -0.04 | 8.3e-01 | Click! |
Activity profile of Rorc_Nr1d1 motif
Sorted Z-values of Rorc_Nr1d1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142215027 | 3.37 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr7_-_142215595 | 2.70 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr7_+_45354512 | 2.19 |
ENSMUST00000080885.12
ENSMUST00000211513.2 ENSMUST00000211357.2 |
Dbp
|
D site albumin promoter binding protein |
| chr1_-_120432669 | 2.16 |
ENSMUST00000027639.8
|
Marco
|
macrophage receptor with collagenous structure |
| chr5_+_90708962 | 1.86 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr16_-_18904240 | 1.41 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr1_-_120432477 | 1.17 |
ENSMUST00000186432.3
|
Marco
|
macrophage receptor with collagenous structure |
| chr3_-_10273628 | 1.13 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr17_+_43671314 | 1.11 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr10_-_128237087 | 1.02 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr14_+_79086492 | 0.96 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr14_+_73379930 | 0.89 |
ENSMUST00000170370.8
ENSMUST00000164822.8 ENSMUST00000165429.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chrX_-_135104589 | 0.84 |
ENSMUST00000066819.11
|
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr17_+_25105617 | 0.82 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr18_-_39000056 | 0.64 |
ENSMUST00000236630.2
ENSMUST00000237356.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr17_+_33857030 | 0.57 |
ENSMUST00000052079.8
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1 |
| chr13_+_6598185 | 0.56 |
ENSMUST00000021611.10
ENSMUST00000222485.2 |
Pitrm1
|
pitrilysin metallepetidase 1 |
| chr11_+_4833186 | 0.55 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr7_-_105249308 | 0.54 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr13_+_52750883 | 0.53 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr4_+_115641996 | 0.49 |
ENSMUST00000177280.8
ENSMUST00000176047.8 |
Atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chrX_-_135104386 | 0.46 |
ENSMUST00000151592.8
ENSMUST00000131510.2 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr11_+_114780867 | 0.46 |
ENSMUST00000106582.9
ENSMUST00000045151.6 |
Cd300a
|
CD300A molecule |
| chr5_+_31454787 | 0.45 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr4_+_94627513 | 0.45 |
ENSMUST00000073939.13
ENSMUST00000102798.8 |
Tek
|
TEK receptor tyrosine kinase |
| chr3_+_108291145 | 0.44 |
ENSMUST00000090561.10
ENSMUST00000102629.8 ENSMUST00000128089.2 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chr2_+_90716204 | 0.44 |
ENSMUST00000111466.3
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr8_+_3715747 | 0.43 |
ENSMUST00000014118.4
|
Mcemp1
|
mast cell expressed membrane protein 1 |
| chr15_-_5137975 | 0.42 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr19_+_6434416 | 0.42 |
ENSMUST00000035269.15
ENSMUST00000113483.2 |
Pygm
|
muscle glycogen phosphorylase |
| chr10_+_25317309 | 0.41 |
ENSMUST00000217929.2
ENSMUST00000220121.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr6_+_29361408 | 0.40 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr3_+_90511068 | 0.38 |
ENSMUST00000001046.7
|
S100a4
|
S100 calcium binding protein A4 |
| chr7_+_45276906 | 0.38 |
ENSMUST00000057927.10
|
Rasip1
|
Ras interacting protein 1 |
| chr15_-_66673425 | 0.37 |
ENSMUST00000168589.8
|
Sla
|
src-like adaptor |
| chr7_+_126810780 | 0.36 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr2_+_155593030 | 0.36 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr15_+_58287305 | 0.36 |
ENSMUST00000037270.5
|
Fam91a1
|
family with sequence similarity 91, member A1 |
| chr17_-_46343291 | 0.35 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr13_-_63036096 | 0.35 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr11_-_110058899 | 0.34 |
ENSMUST00000044850.4
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr8_-_95405234 | 0.34 |
ENSMUST00000213043.2
|
Pllp
|
plasma membrane proteolipid |
| chr15_-_5137951 | 0.34 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr7_+_114367971 | 0.33 |
ENSMUST00000117543.3
ENSMUST00000151464.2 |
Insc
|
INSC spindle orientation adaptor protein |
| chr17_-_46342739 | 0.33 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr9_-_30833748 | 0.31 |
ENSMUST00000065112.7
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr12_-_84923252 | 0.31 |
ENSMUST00000163189.8
ENSMUST00000110254.9 ENSMUST00000002073.13 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr6_-_88851027 | 0.29 |
ENSMUST00000038409.12
|
Podxl2
|
podocalyxin-like 2 |
| chr14_-_60434622 | 0.29 |
ENSMUST00000131670.3
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr3_+_122305819 | 0.27 |
ENSMUST00000199344.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr7_-_133833734 | 0.27 |
ENSMUST00000134504.8
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr4_+_11558905 | 0.26 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr12_-_40298072 | 0.26 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr11_+_87471867 | 0.26 |
ENSMUST00000018544.12
ENSMUST00000063156.11 ENSMUST00000107960.8 |
Septin4
|
septin 4 |
| chr3_+_53371086 | 0.25 |
ENSMUST00000058577.5
|
Proser1
|
proline and serine rich 1 |
| chr3_-_53370621 | 0.25 |
ENSMUST00000056749.14
|
Nhlrc3
|
NHL repeat containing 3 |
| chr11_+_101136821 | 0.24 |
ENSMUST00000129680.8
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr7_-_79115760 | 0.24 |
ENSMUST00000125562.2
|
Polg
|
polymerase (DNA directed), gamma |
| chr12_+_3941728 | 0.24 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr7_+_126895463 | 0.23 |
ENSMUST00000106306.9
ENSMUST00000120857.8 |
Itgal
|
integrin alpha L |
| chr7_-_79115915 | 0.22 |
ENSMUST00000073889.14
|
Polg
|
polymerase (DNA directed), gamma |
| chr5_+_135216090 | 0.22 |
ENSMUST00000002825.6
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr3_-_153650068 | 0.20 |
ENSMUST00000150070.2
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr7_-_99770280 | 0.19 |
ENSMUST00000208184.2
|
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr5_-_122917341 | 0.19 |
ENSMUST00000198257.5
ENSMUST00000199599.2 ENSMUST00000196742.2 ENSMUST00000200109.5 ENSMUST00000111668.8 |
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr3_-_92493507 | 0.19 |
ENSMUST00000194965.6
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr12_+_116011332 | 0.19 |
ENSMUST00000073551.6
ENSMUST00000183125.2 |
Zfp386
|
zinc finger protein 386 (Kruppel-like) |
| chr7_+_126895531 | 0.18 |
ENSMUST00000170971.8
|
Itgal
|
integrin alpha L |
| chr6_-_93890520 | 0.18 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr19_+_31846154 | 0.18 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr10_+_80003612 | 0.17 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein |
| chr15_-_10485976 | 0.17 |
ENSMUST00000169050.8
ENSMUST00000022855.12 |
Brix1
|
BRX1, biogenesis of ribosomes |
| chr7_-_30738471 | 0.17 |
ENSMUST00000162250.8
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr7_+_126895423 | 0.17 |
ENSMUST00000117762.8
|
Itgal
|
integrin alpha L |
| chr18_+_35987791 | 0.17 |
ENSMUST00000235404.2
|
Cxxc5
|
CXXC finger 5 |
| chr14_-_36657517 | 0.16 |
ENSMUST00000183038.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr14_-_40730180 | 0.16 |
ENSMUST00000136661.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr6_-_93890659 | 0.16 |
ENSMUST00000093769.8
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr5_-_65492940 | 0.16 |
ENSMUST00000203471.3
ENSMUST00000172732.8 ENSMUST00000204965.3 |
Rfc1
|
replication factor C (activator 1) 1 |
| chr11_+_95603494 | 0.16 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr2_+_153133326 | 0.15 |
ENSMUST00000028977.7
|
Kif3b
|
kinesin family member 3B |
| chr17_-_45997132 | 0.14 |
ENSMUST00000113523.9
|
Tmem63b
|
transmembrane protein 63b |
| chr2_+_48839505 | 0.14 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr5_-_5564873 | 0.14 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr12_-_72455708 | 0.14 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1 |
| chr9_+_75348800 | 0.14 |
ENSMUST00000048937.6
|
Leo1
|
Leo1, Paf1/RNA polymerase II complex component |
| chr9_+_65368207 | 0.13 |
ENSMUST00000034955.8
ENSMUST00000213957.2 |
Spg21
|
SPG21, maspardin |
| chr4_-_116228921 | 0.13 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr2_+_152596075 | 0.13 |
ENSMUST00000010020.12
|
Cox4i2
|
cytochrome c oxidase subunit 4I2 |
| chr11_-_3599673 | 0.12 |
ENSMUST00000193809.2
|
Tug1
|
taurine upregulated gene 1 |
| chr17_-_34847354 | 0.11 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr14_+_105496147 | 0.11 |
ENSMUST00000138283.2
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr13_+_104365432 | 0.11 |
ENSMUST00000070761.10
ENSMUST00000225557.2 |
Cenpk
|
centromere protein K |
| chr6_-_30873669 | 0.11 |
ENSMUST00000048774.13
ENSMUST00000166192.7 |
Copg2
|
coatomer protein complex, subunit gamma 2 |
| chr17_-_34847013 | 0.10 |
ENSMUST00000166040.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr15_+_10486092 | 0.09 |
ENSMUST00000022856.15
ENSMUST00000100775.10 ENSMUST00000169519.8 |
Rad1
|
RAD1 checkpoint DNA exonuclease |
| chr15_-_11996084 | 0.08 |
ENSMUST00000022816.15
|
Sub1
|
SUB1 homolog, transcriptional regulator |
| chr15_-_98560739 | 0.08 |
ENSMUST00000162384.2
ENSMUST00000003450.15 |
Ddx23
|
DEAD box helicase 23 |
| chr19_-_43928583 | 0.08 |
ENSMUST00000212396.2
|
Dnmbp
|
dynamin binding protein |
| chr7_-_133833854 | 0.08 |
ENSMUST00000127524.2
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
| chr3_-_103716593 | 0.08 |
ENSMUST00000063502.13
ENSMUST00000106832.2 ENSMUST00000106834.8 ENSMUST00000029435.15 |
Dclre1b
|
DNA cross-link repair 1B |
| chr11_+_58531220 | 0.08 |
ENSMUST00000075084.5
|
Trim58
|
tripartite motif-containing 58 |
| chr11_+_116089678 | 0.08 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr11_+_96822213 | 0.08 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr11_+_51989508 | 0.07 |
ENSMUST00000020608.3
|
Ppp2ca
|
protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform |
| chr3_+_79791798 | 0.06 |
ENSMUST00000118853.8
ENSMUST00000145992.2 |
Gask1b
|
golgi associated kinase 1B |
| chr14_-_55101505 | 0.06 |
ENSMUST00000142283.4
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr12_-_34578842 | 0.06 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr7_-_99276310 | 0.05 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr16_-_20549294 | 0.05 |
ENSMUST00000231826.2
ENSMUST00000076422.13 ENSMUST00000232217.2 |
Thpo
|
thrombopoietin |
| chr6_-_124519240 | 0.05 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr5_+_145020910 | 0.05 |
ENSMUST00000124379.3
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr11_-_51526697 | 0.05 |
ENSMUST00000001081.10
|
Rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr11_+_40624763 | 0.05 |
ENSMUST00000127382.2
|
Nudcd2
|
NudC domain containing 2 |
| chr10_+_130158737 | 0.04 |
ENSMUST00000217702.2
ENSMUST00000042586.10 ENSMUST00000218605.2 |
Tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr7_-_126224848 | 0.04 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr5_+_145020640 | 0.04 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr14_+_33775423 | 0.03 |
ENSMUST00000058725.5
|
Antxrl
|
anthrax toxin receptor-like |
| chr5_-_65492907 | 0.03 |
ENSMUST00000203581.3
|
Rfc1
|
replication factor C (activator 1) 1 |
| chr1_+_86631510 | 0.02 |
ENSMUST00000168237.8
ENSMUST00000065694.8 |
Dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr16_+_21828223 | 0.02 |
ENSMUST00000023561.8
|
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chrX_-_37665041 | 0.02 |
ENSMUST00000050083.6
ENSMUST00000115118.8 |
Cul4b
|
cullin 4B |
| chr14_+_53100756 | 0.02 |
ENSMUST00000103616.5
ENSMUST00000186370.2 |
Trav15d-1-dv6d-1
|
T cell receptor alpha variable 15D-1-DV6D-1 |
| chr11_-_4045343 | 0.02 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr7_-_119461027 | 0.02 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr7_+_45683122 | 0.02 |
ENSMUST00000033121.7
|
Nomo1
|
nodal modulator 1 |
| chr2_+_96148418 | 0.01 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr16_-_44379226 | 0.01 |
ENSMUST00000114634.3
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr2_-_152857239 | 0.01 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr11_-_99176086 | 0.00 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
| chr1_-_131161312 | 0.00 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr6_+_71176811 | 0.00 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rorc_Nr1d1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.7 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.2 | 1.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 0.5 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of eosinophil activation(GO:1902567) negative regulation of activation of JAK2 kinase activity(GO:1902569) negative regulation of eosinophil migration(GO:2000417) |
| 0.1 | 0.5 | GO:0071226 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.1 | 0.5 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 3.2 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 1.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.1 | 0.5 | GO:0060332 | heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.6 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.5 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 1.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.6 | GO:1902563 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.1 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.3 | GO:0061091 | involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.5 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.4 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 0.6 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 3.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 6.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 3.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.2 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 6.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |