Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
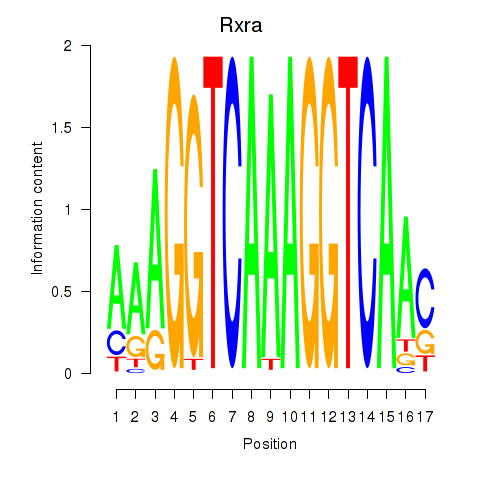
Results for Rxra
Z-value: 1.22
Transcription factors associated with Rxra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxra
|
ENSMUSG00000015846.16 | retinoid X receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxra | mm39_v1_chr2_+_27567213_27567241 | 0.59 | 1.7e-04 | Click! |
Activity profile of Rxra motif
Sorted Z-values of Rxra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39275518 | 28.65 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr19_+_20470056 | 16.30 |
ENSMUST00000225337.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr7_+_26821266 | 15.08 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr3_+_138121245 | 13.47 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr19_+_20470114 | 13.19 |
ENSMUST00000225313.2
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr2_+_172994841 | 11.12 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr11_-_116089866 | 10.53 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr15_+_82336535 | 10.53 |
ENSMUST00000089129.7
ENSMUST00000229313.2 ENSMUST00000231136.2 |
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr15_-_82264379 | 8.28 |
ENSMUST00000023083.9
|
Cyp2d22
|
cytochrome P450, family 2, subfamily d, polypeptide 22 |
| chr6_-_85797946 | 8.09 |
ENSMUST00000032074.5
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr11_-_78313043 | 7.72 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr13_-_24098981 | 7.15 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr18_+_12732951 | 7.00 |
ENSMUST00000234255.2
ENSMUST00000169401.8 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr13_-_24098951 | 6.60 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr15_-_100579450 | 6.58 |
ENSMUST00000230740.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr11_-_116089595 | 6.53 |
ENSMUST00000072948.11
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr12_-_103796632 | 6.37 |
ENSMUST00000164454.3
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr15_-_100579813 | 6.24 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr1_+_74324089 | 5.84 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr12_-_103706774 | 5.80 |
ENSMUST00000186166.7
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr1_-_180073492 | 5.78 |
ENSMUST00000010753.14
|
Psen2
|
presenilin 2 |
| chr9_-_103165489 | 5.68 |
ENSMUST00000035163.10
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr9_-_103165423 | 5.59 |
ENSMUST00000123530.8
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr17_-_74257164 | 4.68 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr5_-_87402659 | 4.45 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr2_+_155223728 | 4.42 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr1_-_180073322 | 4.10 |
ENSMUST00000111104.2
|
Psen2
|
presenilin 2 |
| chr19_-_44017637 | 3.53 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr5_+_92719336 | 3.38 |
ENSMUST00000176621.8
ENSMUST00000175974.2 ENSMUST00000131166.9 ENSMUST00000176448.8 ENSMUST00000082382.8 |
Fam47e
|
family with sequence similarity 47, member E |
| chr2_-_121786573 | 3.35 |
ENSMUST00000104936.4
|
Mageb3
|
MAGE family member B3 |
| chr19_-_6899121 | 3.29 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr9_-_103105638 | 3.23 |
ENSMUST00000126359.2
|
Trf
|
transferrin |
| chr4_-_103072343 | 3.18 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr11_+_114566257 | 3.09 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr19_+_36532061 | 2.98 |
ENSMUST00000169036.9
ENSMUST00000047247.12 |
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr19_-_6899173 | 2.93 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr19_-_3962733 | 2.80 |
ENSMUST00000075092.8
ENSMUST00000235847.2 ENSMUST00000235301.2 ENSMUST00000237341.2 |
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr14_+_69409251 | 2.69 |
ENSMUST00000062437.10
|
Nkx2-6
|
NK2 homeobox 6 |
| chr17_-_57024660 | 2.65 |
ENSMUST00000164907.3
ENSMUST00000233208.2 |
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr6_+_133082202 | 2.58 |
ENSMUST00000191462.2
|
Smim10l1
|
small integral membrane protein 10 like 1 |
| chr6_+_78402956 | 2.57 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr2_+_91033230 | 2.41 |
ENSMUST00000150403.8
ENSMUST00000002172.14 ENSMUST00000238832.2 ENSMUST00000239169.2 ENSMUST00000155418.2 |
Acp2
|
acid phosphatase 2, lysosomal |
| chr16_+_4825216 | 2.25 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr10_-_7656635 | 2.02 |
ENSMUST00000124838.2
ENSMUST00000039763.14 |
Ginm1
|
glycoprotein integral membrane 1 |
| chr9_+_65538352 | 2.01 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr19_-_21449916 | 1.94 |
ENSMUST00000087600.10
|
Gda
|
guanine deaminase |
| chr2_-_69172944 | 1.89 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr10_+_77417554 | 1.87 |
ENSMUST00000009435.12
|
Pttg1ip
|
pituitary tumor-transforming 1 interacting protein |
| chr10_+_62897353 | 1.86 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr10_+_77417608 | 1.85 |
ENSMUST00000162429.8
|
Pttg1ip
|
pituitary tumor-transforming 1 interacting protein |
| chr16_+_4825146 | 1.82 |
ENSMUST00000184439.8
|
Smim22
|
small integral membrane protein 22 |
| chr1_-_63215952 | 1.78 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr9_-_44876817 | 1.76 |
ENSMUST00000214761.2
ENSMUST00000213666.2 ENSMUST00000213890.2 ENSMUST00000125642.8 ENSMUST00000213193.2 ENSMUST00000117506.9 ENSMUST00000138559.9 ENSMUST00000117549.8 |
Ube4a
|
ubiquitination factor E4A |
| chr11_-_53313950 | 1.74 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr17_+_57024759 | 1.72 |
ENSMUST00000002452.8
ENSMUST00000233832.2 ENSMUST00000233233.2 |
Ndufa11
|
NADH:ubiquinone oxidoreductase subunit A11 |
| chr19_+_5928649 | 1.67 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr16_+_4825170 | 1.65 |
ENSMUST00000178155.9
|
Smim22
|
small integral membrane protein 22 |
| chr4_+_97665843 | 1.63 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr11_+_50917831 | 1.57 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr4_+_97665992 | 1.56 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr17_-_28039506 | 1.56 |
ENSMUST00000114859.9
ENSMUST00000233533.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr2_-_119060330 | 1.52 |
ENSMUST00000110820.3
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr14_+_122116002 | 1.52 |
ENSMUST00000039803.7
|
Ubac2
|
ubiquitin associated domain containing 2 |
| chr9_+_44684248 | 1.49 |
ENSMUST00000128150.8
|
Ift46
|
intraflagellar transport 46 |
| chr6_+_73225616 | 1.47 |
ENSMUST00000203632.2
|
Suclg1
|
succinate-CoA ligase, GDP-forming, alpha subunit |
| chr8_-_71085097 | 1.47 |
ENSMUST00000110103.2
|
Gdf15
|
growth differentiation factor 15 |
| chr17_-_15784582 | 1.46 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chr6_-_85797917 | 1.38 |
ENSMUST00000174143.2
|
Nat8f6
|
N-acetyltransferase 8 (GCN5-related) family member 6 |
| chr4_+_126450762 | 1.38 |
ENSMUST00000147675.2
|
Clspn
|
claspin |
| chr1_+_75456173 | 1.38 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr3_+_100732768 | 1.37 |
ENSMUST00000054791.9
|
Vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr14_+_12016304 | 1.35 |
ENSMUST00000161302.8
|
Fhit
|
fragile histidine triad gene |
| chr10_+_77418221 | 1.34 |
ENSMUST00000162943.9
|
Pttg1ip
|
pituitary tumor-transforming 1 interacting protein |
| chr10_-_67384898 | 1.32 |
ENSMUST00000075686.7
|
Ado
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr9_+_44684206 | 1.29 |
ENSMUST00000002099.11
|
Ift46
|
intraflagellar transport 46 |
| chr12_-_79239022 | 1.27 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr6_+_41498716 | 1.27 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr2_-_3420102 | 1.26 |
ENSMUST00000115082.10
|
Meig1
|
meiosis expressed gene 1 |
| chr8_-_100143029 | 1.26 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr9_+_44684285 | 1.25 |
ENSMUST00000239014.2
|
Ift46
|
intraflagellar transport 46 |
| chr1_-_63215812 | 1.22 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr7_-_98305986 | 1.19 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr9_+_108269992 | 1.19 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr2_-_3420056 | 1.17 |
ENSMUST00000115084.8
ENSMUST00000115083.8 |
Meig1
|
meiosis expressed gene 1 |
| chr19_+_10502679 | 1.14 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr10_+_23836392 | 1.09 |
ENSMUST00000092660.2
|
Taar4
|
trace amine-associated receptor 4 |
| chr7_+_132460954 | 1.03 |
ENSMUST00000084497.12
ENSMUST00000106161.8 |
Abraxas2
|
BRISC complex subunit |
| chr16_+_29028860 | 1.02 |
ENSMUST00000162747.8
|
Plaat1
|
phospholipase A and acyltransferase 1 |
| chr17_-_28039588 | 0.98 |
ENSMUST00000114863.10
ENSMUST00000233131.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr6_+_8259328 | 0.98 |
ENSMUST00000159378.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr17_-_56933872 | 0.95 |
ENSMUST00000047226.10
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr7_+_23969822 | 0.95 |
ENSMUST00000108438.10
|
Zfp93
|
zinc finger protein 93 |
| chr6_+_8259379 | 0.94 |
ENSMUST00000162034.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr5_+_14075281 | 0.94 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr1_-_172085977 | 0.93 |
ENSMUST00000111243.2
|
Atp1a4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr9_+_44684450 | 0.92 |
ENSMUST00000238800.2
ENSMUST00000147559.8 |
Ift46
|
intraflagellar transport 46 |
| chr6_+_8259405 | 0.92 |
ENSMUST00000160705.8
ENSMUST00000159433.8 |
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr7_-_29931612 | 0.91 |
ENSMUST00000006254.6
|
Tbcb
|
tubulin folding cofactor B |
| chr2_-_119060366 | 0.91 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr14_+_54014387 | 0.90 |
ENSMUST00000103670.4
|
Trav3-4
|
T cell receptor alpha variable 3-4 |
| chr3_-_120965327 | 0.89 |
ENSMUST00000170781.2
ENSMUST00000039761.12 ENSMUST00000106467.8 ENSMUST00000106466.10 ENSMUST00000164925.9 |
Rwdd3
|
RWD domain containing 3 |
| chr10_-_79582387 | 0.88 |
ENSMUST00000020580.13
ENSMUST00000159016.8 |
Polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr1_+_139429430 | 0.88 |
ENSMUST00000027615.7
|
F13b
|
coagulation factor XIII, beta subunit |
| chr9_+_108270020 | 0.88 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr4_+_118286898 | 0.88 |
ENSMUST00000067896.4
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr4_-_137301880 | 0.87 |
ENSMUST00000178923.2
|
Ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr17_-_36220518 | 0.87 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr4_+_118266582 | 0.87 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr7_+_43093507 | 0.85 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr9_+_44684324 | 0.83 |
ENSMUST00000214854.3
ENSMUST00000125877.8 |
Ift46
|
intraflagellar transport 46 |
| chr15_-_95426419 | 0.81 |
ENSMUST00000229933.2
ENSMUST00000166170.9 |
Nell2
|
NEL-like 2 |
| chr4_+_85123654 | 0.81 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr5_-_129916283 | 0.78 |
ENSMUST00000094280.4
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr2_+_164174660 | 0.78 |
ENSMUST00000017148.8
|
Svs5
|
seminal vesicle secretory protein 5 |
| chr7_-_28913382 | 0.77 |
ENSMUST00000169143.8
ENSMUST00000047846.13 |
Catsperg1
|
cation channel sperm associated auxiliary subunit gamma 1 |
| chr10_+_14581325 | 0.77 |
ENSMUST00000191238.7
ENSMUST00000190114.2 |
Nmbr
|
neuromedin B receptor |
| chr4_-_118266416 | 0.76 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr10_+_74896383 | 0.76 |
ENSMUST00000164107.3
|
Bcr
|
BCR activator of RhoGEF and GTPase |
| chr2_+_58991182 | 0.76 |
ENSMUST00000168631.8
ENSMUST00000102754.11 ENSMUST00000123908.8 |
Pkp4
|
plakophilin 4 |
| chr17_-_36220924 | 0.74 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr7_-_98305737 | 0.74 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chrX_+_158197568 | 0.73 |
ENSMUST00000112471.9
|
Map7d2
|
MAP7 domain containing 2 |
| chr15_-_97902576 | 0.72 |
ENSMUST00000023123.15
|
Col2a1
|
collagen, type II, alpha 1 |
| chr8_+_12997948 | 0.72 |
ENSMUST00000110871.8
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr3_+_27371206 | 0.72 |
ENSMUST00000174840.2
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr13_+_113063988 | 0.71 |
ENSMUST00000038574.7
|
Dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chrX_-_147398149 | 0.71 |
ENSMUST00000112755.3
|
Gm8334
|
predicted gene 8334 |
| chr10_+_78410180 | 0.69 |
ENSMUST00000218061.2
ENSMUST00000218787.2 ENSMUST00000105384.5 ENSMUST00000218875.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr16_+_11140740 | 0.68 |
ENSMUST00000180792.8
|
Snx29
|
sorting nexin 29 |
| chr11_+_97689819 | 0.68 |
ENSMUST00000143571.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr18_+_70605476 | 0.67 |
ENSMUST00000114959.9
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr4_+_118266526 | 0.66 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr16_+_15135322 | 0.65 |
ENSMUST00000178312.2
|
Gm21897
|
predicted gene, 21897 |
| chr3_+_27371168 | 0.65 |
ENSMUST00000046383.12
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr11_-_11577824 | 0.64 |
ENSMUST00000081896.5
|
4930512M02Rik
|
RIKEN cDNA 4930512M02 gene |
| chr12_-_74363168 | 0.64 |
ENSMUST00000110441.2
|
Gm11042
|
predicted gene 11042 |
| chr2_-_32665596 | 0.63 |
ENSMUST00000161430.8
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr11_-_69906171 | 0.62 |
ENSMUST00000018718.8
ENSMUST00000102574.10 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr15_-_97902515 | 0.62 |
ENSMUST00000088355.12
|
Col2a1
|
collagen, type II, alpha 1 |
| chr3_-_87965757 | 0.61 |
ENSMUST00000029708.8
|
Naxe
|
NAD(P)HX epimerase |
| chr11_+_4570067 | 0.61 |
ENSMUST00000109941.2
|
Gm11032
|
predicted gene 11032 |
| chr16_+_11140779 | 0.61 |
ENSMUST00000115814.4
|
Snx29
|
sorting nexin 29 |
| chr2_-_52225763 | 0.59 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr14_-_14512458 | 0.59 |
ENSMUST00000211339.2
|
Gm45521
|
predicted gene 45521 |
| chr2_-_26127360 | 0.59 |
ENSMUST00000036187.9
|
Qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr11_+_109376432 | 0.59 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr14_+_73898665 | 0.56 |
ENSMUST00000098874.4
|
Gm21750
|
predicted gene, 21750 |
| chr3_-_153610528 | 0.56 |
ENSMUST00000190449.2
|
Msh4
|
mutS homolog 4 |
| chr7_+_141048191 | 0.55 |
ENSMUST00000211071.2
|
Cd151
|
CD151 antigen |
| chr4_-_150087587 | 0.54 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr19_-_12302465 | 0.54 |
ENSMUST00000207241.3
|
Olfr1437
|
olfactory receptor 1437 |
| chr7_-_29869126 | 0.54 |
ENSMUST00000062181.9
|
Zfp146
|
zinc finger protein 146 |
| chr11_-_33463627 | 0.53 |
ENSMUST00000037522.14
|
Ranbp17
|
RAN binding protein 17 |
| chr3_-_153610368 | 0.53 |
ENSMUST00000188338.7
|
Msh4
|
mutS homolog 4 |
| chr5_-_24760401 | 0.52 |
ENSMUST00000088302.10
|
Iqca1l
|
IQ motif containing with AAA domain 1 like |
| chr2_-_164041997 | 0.52 |
ENSMUST00000063251.3
|
Wfdc15a
|
WAP four-disulfide core domain 15A |
| chr14_+_53210216 | 0.50 |
ENSMUST00000103599.3
|
Trav3d-3
|
T cell receptor alpha variable 3D-3 |
| chr5_+_31107390 | 0.50 |
ENSMUST00000006814.9
|
Abhd1
|
abhydrolase domain containing 1 |
| chr14_+_53903370 | 0.48 |
ENSMUST00000181768.3
|
Trav3-3
|
T cell receptor alpha variable 3-3 |
| chr9_-_78285942 | 0.48 |
ENSMUST00000034900.8
|
Ooep
|
oocyte expressed protein |
| chr2_-_32665637 | 0.47 |
ENSMUST00000161958.2
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr17_+_49922129 | 0.45 |
ENSMUST00000162854.2
|
Kif6
|
kinesin family member 6 |
| chr2_-_165210622 | 0.44 |
ENSMUST00000141140.2
ENSMUST00000103085.8 |
Zfp663
|
zinc finger protein 663 |
| chr8_-_85741037 | 0.43 |
ENSMUST00000059072.6
ENSMUST00000209421.2 |
Best2
|
bestrophin 2 |
| chr1_-_84912810 | 0.43 |
ENSMUST00000027422.7
|
Slc16a14
|
solute carrier family 16 (monocarboxylic acid transporters), member 14 |
| chr17_-_57025022 | 0.43 |
ENSMUST00000233400.2
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr9_+_45014092 | 0.43 |
ENSMUST00000217074.2
|
Jaml
|
junction adhesion molecule like |
| chr5_-_115257336 | 0.42 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr7_-_43906629 | 0.41 |
ENSMUST00000012921.9
|
Acp4
|
acid phosphatase 4 |
| chr5_-_109704339 | 0.41 |
ENSMUST00000198960.2
|
Crlf2
|
cytokine receptor-like factor 2 |
| chr1_+_165957909 | 0.41 |
ENSMUST00000166159.2
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr8_-_4267459 | 0.40 |
ENSMUST00000176227.2
|
Prr36
|
proline rich 36 |
| chr11_+_5578738 | 0.40 |
ENSMUST00000137933.2
|
Ankrd36
|
ankyrin repeat domain 36 |
| chrX_+_6327790 | 0.40 |
ENSMUST00000143641.4
|
Shroom4
|
shroom family member 4 |
| chr12_-_33142858 | 0.40 |
ENSMUST00000095774.3
|
Cdhr3
|
cadherin-related family member 3 |
| chrX_+_99537897 | 0.39 |
ENSMUST00000033570.6
|
Igbp1
|
immunoglobulin (CD79A) binding protein 1 |
| chr5_-_73132045 | 0.39 |
ENSMUST00000043711.9
|
Gm10135
|
predicted gene 10135 |
| chr1_-_57445576 | 0.38 |
ENSMUST00000160837.8
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr4_+_118560720 | 0.37 |
ENSMUST00000214131.2
ENSMUST00000215117.3 |
Olfr1341
|
olfactory receptor 1341 |
| chr5_-_143831842 | 0.36 |
ENSMUST00000079624.12
ENSMUST00000110717.9 |
Ankrd61
|
ankyrin repeat domain 61 |
| chr1_-_74323795 | 0.35 |
ENSMUST00000178235.8
ENSMUST00000006462.14 |
Aamp
|
angio-associated migratory protein |
| chrX_-_41000746 | 0.34 |
ENSMUST00000047037.15
|
Thoc2
|
THO complex 2 |
| chr8_-_46452896 | 0.34 |
ENSMUST00000053558.10
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr2_+_126057020 | 0.33 |
ENSMUST00000164042.3
|
Gm17555
|
predicted gene, 17555 |
| chr2_+_164158651 | 0.33 |
ENSMUST00000017144.3
|
Svs6
|
seminal vesicle secretory protein 6 |
| chr6_+_21986445 | 0.33 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_+_157870653 | 0.32 |
ENSMUST00000152452.8
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr2_-_21210151 | 0.32 |
ENSMUST00000027992.3
|
Enkur
|
enkurin, TRPC channel interacting protein |
| chr9_-_20984790 | 0.32 |
ENSMUST00000010348.7
|
Fdx2
|
ferredoxin 2 |
| chr17_-_35069136 | 0.32 |
ENSMUST00000046022.16
|
Skiv2l
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr2_-_129541753 | 0.31 |
ENSMUST00000028883.12
|
Pdyn
|
prodynorphin |
| chr19_+_4008645 | 0.30 |
ENSMUST00000179433.8
|
Aldh3b3
|
aldehyde dehydrogenase 3 family, member B3 |
| chr2_+_121786444 | 0.29 |
ENSMUST00000036647.13
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr7_-_4550523 | 0.29 |
ENSMUST00000206023.2
|
Syt5
|
synaptotagmin V |
| chr11_-_106163753 | 0.28 |
ENSMUST00000021052.16
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr2_-_19558719 | 0.28 |
ENSMUST00000062060.5
|
4921504E06Rik
|
RIKEN cDNA 4921504E06 gene |
| chr11_-_53509485 | 0.28 |
ENSMUST00000000889.7
|
Il4
|
interleukin 4 |
| chr2_+_88644840 | 0.28 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr15_-_77330396 | 0.26 |
ENSMUST00000229434.2
|
Apol7b
|
apolipoprotein L 7b |
| chr13_+_33668761 | 0.26 |
ENSMUST00000081927.4
|
Serpinb9g
|
serine (or cysteine) peptidase inhibitor, clade B, member 9g |
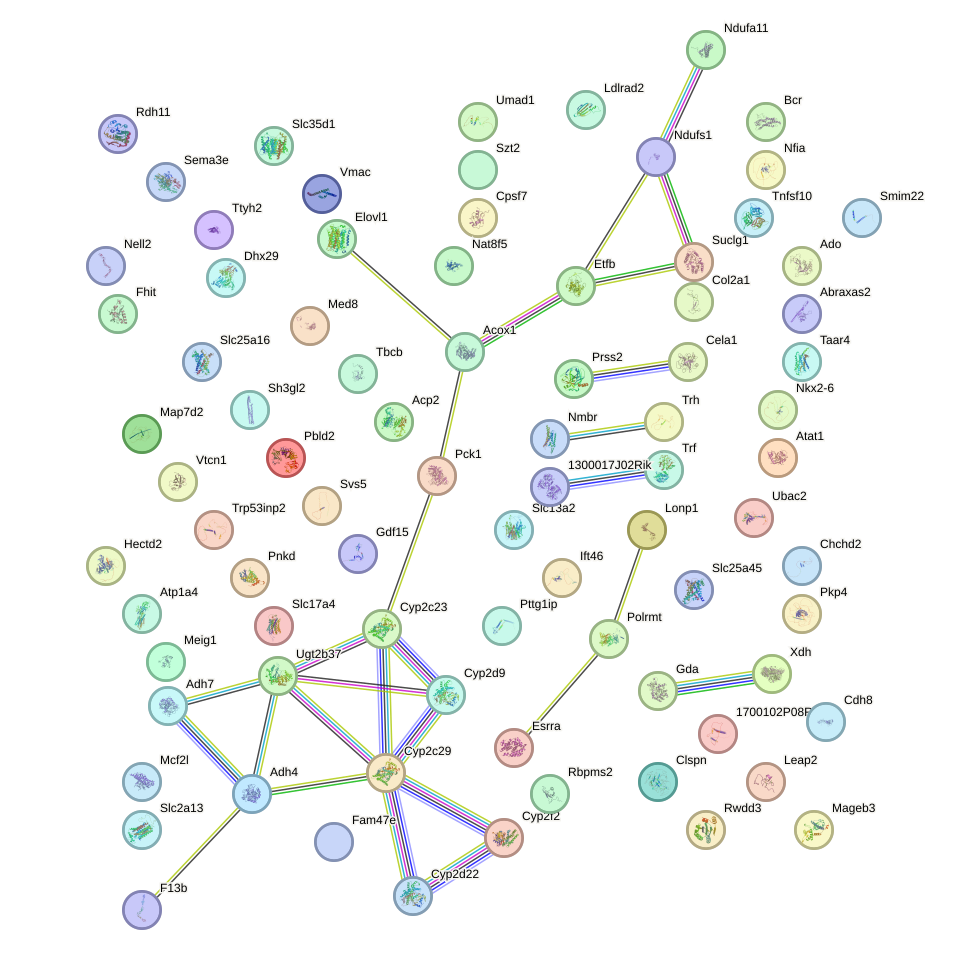
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:0018931 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 3.4 | 13.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 3.3 | 29.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 2.8 | 17.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 2.8 | 11.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.8 | 12.8 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 1.0 | 8.3 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.9 | 4.6 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.9 | 28.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.7 | 2.7 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.6 | 1.9 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.6 | 9.9 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.6 | 5.8 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.5 | 1.6 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.5 | 1.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.5 | 1.9 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.4 | 3.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.3 | 3.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 1.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 0.9 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.3 | 10.5 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 1.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 5.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 12.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 5.8 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 2.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 2.4 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 3.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 1.4 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 1.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 7.9 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 1.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.4 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.4 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 1.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 2.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.8 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.4 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 6.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 1.0 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.5 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 0.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 4.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 13.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 1.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.9 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.3 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.1 | 1.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 8.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 1.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.8 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 1.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 4.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 3.0 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 1.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.4 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 1.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.0 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 3.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.9 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.6 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.5 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 3.2 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 3.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 0.9 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 1.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 1.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.2 | 16.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 5.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 7.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 2.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 3.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 66.4 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 29.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 4.8 | 28.7 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 4.5 | 13.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 3.7 | 11.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 2.4 | 17.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 1.0 | 9.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.9 | 4.7 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.9 | 13.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.8 | 25.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.7 | 5.8 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.5 | 1.5 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.5 | 3.2 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.4 | 8.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) |
| 0.4 | 3.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.3 | 1.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 1.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 1.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.6 | GO:0017099 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.2 | 5.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 0.5 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.2 | 2.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 3.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 4.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 12.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 6.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 6.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 5.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 8.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 6.9 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 8.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.6 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 3.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 4.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 1.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 9.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 15.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 43.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.7 | 17.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.6 | 9.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.5 | 9.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.5 | 6.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.5 | 7.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 3.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 1.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 8.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 6.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |