Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
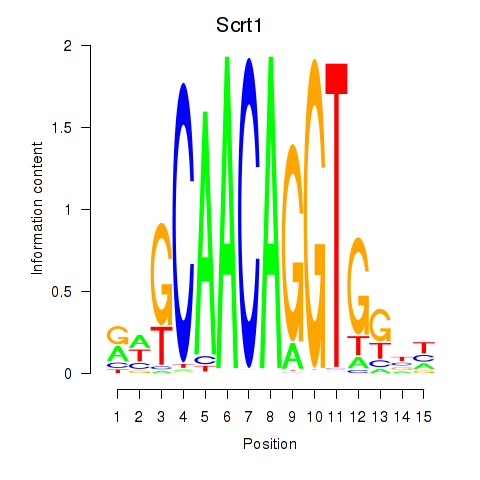
Results for Scrt1
Z-value: 1.09
Transcription factors associated with Scrt1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt1
|
ENSMUSG00000048385.10 | scratch family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt1 | mm39_v1_chr15_-_76406102_76406258 | -0.52 | 1.0e-03 | Click! |
Activity profile of Scrt1 motif
Sorted Z-values of Scrt1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_30623592 | 14.10 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr19_-_40062174 | 12.48 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr10_-_127843377 | 9.25 |
ENSMUST00000219447.2
ENSMUST00000219780.2 ENSMUST00000219707.2 ENSMUST00000219953.2 ENSMUST00000219183.2 |
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr7_+_140343652 | 8.26 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr1_-_150341911 | 7.98 |
ENSMUST00000162367.8
ENSMUST00000161611.8 ENSMUST00000161320.8 ENSMUST00000159035.2 |
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
| chr11_+_83637766 | 6.52 |
ENSMUST00000070832.3
|
Wfdc21
|
WAP four-disulfide core domain 21 |
| chr10_-_81127057 | 6.05 |
ENSMUST00000045744.7
|
Tjp3
|
tight junction protein 3 |
| chr8_-_71990085 | 6.02 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr7_-_30672747 | 5.83 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr7_-_30672824 | 5.57 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr10_-_81127334 | 5.50 |
ENSMUST00000219479.2
|
Tjp3
|
tight junction protein 3 |
| chr6_+_138117519 | 5.48 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr7_-_30672889 | 5.45 |
ENSMUST00000001279.15
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr6_+_138118565 | 5.21 |
ENSMUST00000118091.8
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr9_+_78164402 | 4.44 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr15_-_100576715 | 4.41 |
ENSMUST00000229869.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr7_-_105249308 | 4.27 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr13_-_53135064 | 3.94 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr12_-_103623418 | 3.91 |
ENSMUST00000044159.7
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr12_+_87193922 | 3.59 |
ENSMUST00000222885.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr18_+_56565188 | 3.34 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr10_-_106959444 | 3.33 |
ENSMUST00000165067.9
|
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr3_+_123061094 | 3.32 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr8_-_85620537 | 3.30 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr12_-_103871146 | 3.28 |
ENSMUST00000074051.6
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr4_-_57916283 | 3.24 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr9_-_121745354 | 3.20 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr11_-_59340739 | 3.13 |
ENSMUST00000136436.2
ENSMUST00000150297.2 ENSMUST00000010038.10 ENSMUST00000156146.8 ENSMUST00000132969.8 ENSMUST00000120940.8 |
Snap47
|
synaptosomal-associated protein, 47 |
| chr11_-_12414947 | 3.12 |
ENSMUST00000046755.14
ENSMUST00000109651.9 |
Cobl
|
cordon-bleu WH2 repeat |
| chr15_-_96947963 | 3.07 |
ENSMUST00000230907.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr17_-_56312555 | 2.78 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr11_-_115078653 | 2.75 |
ENSMUST00000103041.8
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr10_+_75768964 | 2.73 |
ENSMUST00000219839.2
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr18_-_3281089 | 2.69 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr19_+_37425180 | 2.68 |
ENSMUST00000128184.3
|
Hhex
|
hematopoietically expressed homeobox |
| chr10_+_116137277 | 2.66 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr5_+_138170259 | 2.65 |
ENSMUST00000019662.11
ENSMUST00000151318.8 |
Ap4m1
|
adaptor-related protein complex AP-4, mu 1 |
| chr9_-_110709175 | 2.53 |
ENSMUST00000050958.9
|
Tmie
|
transmembrane inner ear |
| chr17_+_56312672 | 2.49 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr3_-_89009214 | 2.49 |
ENSMUST00000081848.13
|
Fdps
|
farnesyl diphosphate synthetase |
| chr19_+_11514132 | 2.46 |
ENSMUST00000025581.7
|
Ms4a4d
|
membrane-spanning 4-domains, subfamily A, member 4D |
| chr3_-_89009153 | 2.46 |
ENSMUST00000199668.3
ENSMUST00000196709.5 |
Fdps
|
farnesyl diphosphate synthetase |
| chr7_-_127545896 | 2.40 |
ENSMUST00000118755.8
ENSMUST00000094026.10 |
Prss36
|
protease, serine 36 |
| chr11_-_12414850 | 2.34 |
ENSMUST00000109650.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr10_+_128158413 | 2.34 |
ENSMUST00000219836.2
|
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr1_-_183150867 | 2.30 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chr12_-_103923145 | 2.29 |
ENSMUST00000085054.5
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr2_+_24866039 | 2.28 |
ENSMUST00000045295.14
|
Pnpla7
|
patatin-like phospholipase domain containing 7 |
| chr10_-_106959462 | 2.28 |
ENSMUST00000044668.5
|
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr6_-_23839136 | 2.21 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr15_-_77813123 | 2.20 |
ENSMUST00000109748.9
ENSMUST00000109747.9 ENSMUST00000100486.6 ENSMUST00000005487.12 |
Txn2
|
thioredoxin 2 |
| chr6_+_78402956 | 2.15 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr5_+_137568113 | 2.14 |
ENSMUST00000031729.13
ENSMUST00000199054.5 |
Tfr2
|
transferrin receptor 2 |
| chr8_+_77628916 | 2.14 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr5_+_137568086 | 2.12 |
ENSMUST00000198866.5
|
Tfr2
|
transferrin receptor 2 |
| chr1_-_155912159 | 2.10 |
ENSMUST00000097527.10
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr5_-_33815760 | 2.09 |
ENSMUST00000019439.9
|
Tmem129
|
transmembrane protein 129 |
| chr10_+_128158328 | 2.00 |
ENSMUST00000219037.2
ENSMUST00000026446.4 |
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr12_-_86931529 | 1.97 |
ENSMUST00000038422.8
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr7_-_133378410 | 1.97 |
ENSMUST00000130182.2
ENSMUST00000106139.8 |
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr18_-_3281727 | 1.88 |
ENSMUST00000154705.8
ENSMUST00000151084.8 |
Crem
|
cAMP responsive element modulator |
| chr1_+_131794962 | 1.87 |
ENSMUST00000112386.8
ENSMUST00000027693.8 |
Rab29
|
RAB29, member RAS oncogene family |
| chr7_+_30673212 | 1.81 |
ENSMUST00000129773.2
|
Fam187b
|
family with sequence similarity 187, member B |
| chr2_+_153334710 | 1.81 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr7_-_88987917 | 1.78 |
ENSMUST00000137723.2
ENSMUST00000117852.8 ENSMUST00000041968.11 |
Tmem135
|
transmembrane protein 135 |
| chr11_-_12414804 | 1.78 |
ENSMUST00000172919.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr9_+_107765320 | 1.77 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr16_+_3702604 | 1.76 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr16_+_3702523 | 1.75 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr7_-_44318710 | 1.73 |
ENSMUST00000208131.2
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr15_-_75886166 | 1.70 |
ENSMUST00000060807.12
|
Fam83h
|
family with sequence similarity 83, member H |
| chr16_+_16888084 | 1.68 |
ENSMUST00000231514.2
|
Ypel1
|
yippee like 1 |
| chr16_+_16887991 | 1.64 |
ENSMUST00000232258.2
|
Ypel1
|
yippee like 1 |
| chr8_-_106863423 | 1.63 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr4_-_129083251 | 1.60 |
ENSMUST00000117965.8
|
S100pbp
|
S100P binding protein |
| chr14_+_52122439 | 1.60 |
ENSMUST00000167984.2
|
Mettl17
|
methyltransferase like 17 |
| chr17_-_56490887 | 1.58 |
ENSMUST00000019723.8
|
Mydgf
|
myeloid derived growth factor |
| chr7_-_44465998 | 1.56 |
ENSMUST00000209072.2
ENSMUST00000047356.11 |
Atf5
|
activating transcription factor 5 |
| chr7_-_133378468 | 1.56 |
ENSMUST00000033290.12
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr11_+_100960838 | 1.55 |
ENSMUST00000001802.10
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chr16_+_36514386 | 1.54 |
ENSMUST00000119464.2
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr19_+_24976864 | 1.53 |
ENSMUST00000025831.8
|
Dock8
|
dedicator of cytokinesis 8 |
| chr16_+_36514334 | 1.51 |
ENSMUST00000023617.13
ENSMUST00000089618.10 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr6_-_148732893 | 1.50 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr7_-_133384449 | 1.49 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chrX_+_5959507 | 1.49 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr8_-_106863521 | 1.48 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr19_-_21630143 | 1.48 |
ENSMUST00000179768.8
ENSMUST00000178523.2 ENSMUST00000038830.10 |
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr13_-_34837469 | 1.47 |
ENSMUST00000053459.15
|
Pxdc1
|
PX domain containing 1 |
| chr6_-_90693471 | 1.47 |
ENSMUST00000101153.10
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr1_-_156131155 | 1.44 |
ENSMUST00000141760.4
ENSMUST00000121146.10 |
Tdrd5
|
tudor domain containing 5 |
| chr16_+_16888145 | 1.42 |
ENSMUST00000232574.2
|
Ypel1
|
yippee like 1 |
| chr7_+_99659121 | 1.39 |
ENSMUST00000107084.8
|
Chrdl2
|
chordin-like 2 |
| chr17_-_25300112 | 1.37 |
ENSMUST00000024984.7
|
Tmem204
|
transmembrane protein 204 |
| chr16_-_4950285 | 1.37 |
ENSMUST00000035672.5
|
Ppl
|
periplakin |
| chr10_-_102866076 | 1.36 |
ENSMUST00000218282.2
ENSMUST00000170026.2 |
Alx1
Gm17028
|
ALX homeobox 1 predicted gene 17028 |
| chr4_-_135221810 | 1.35 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr1_-_189075903 | 1.32 |
ENSMUST00000192723.2
ENSMUST00000110920.7 |
Kcnk2
|
potassium channel, subfamily K, member 2 |
| chr4_-_83242366 | 1.31 |
ENSMUST00000030205.14
ENSMUST00000048274.11 |
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr6_-_41012435 | 1.30 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr18_-_3280999 | 1.29 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr12_+_4642629 | 1.29 |
ENSMUST00000218402.2
|
Itsn2
|
intersectin 2 |
| chr5_+_37403098 | 1.28 |
ENSMUST00000031004.11
|
Crmp1
|
collapsin response mediator protein 1 |
| chr14_+_55120777 | 1.26 |
ENSMUST00000022806.10
|
Bcl2l2
|
BCL2-like 2 |
| chr18_+_36693024 | 1.26 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr1_+_165288606 | 1.25 |
ENSMUST00000027853.6
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr10_+_62935430 | 1.24 |
ENSMUST00000044059.5
|
Atoh7
|
atonal bHLH transcription factor 7 |
| chr6_-_148732946 | 1.24 |
ENSMUST00000048418.14
|
Ipo8
|
importin 8 |
| chr2_+_131075965 | 1.24 |
ENSMUST00000041362.12
ENSMUST00000110199.3 |
Mavs
|
mitochondrial antiviral signaling protein |
| chr4_-_106474429 | 1.23 |
ENSMUST00000189032.7
ENSMUST00000106788.2 |
Lexm
|
lymphocyte expansion molecule |
| chr17_-_26063488 | 1.23 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr7_-_28981335 | 1.22 |
ENSMUST00000108236.5
ENSMUST00000098604.12 |
Spint2
|
serine protease inhibitor, Kunitz type 2 |
| chr19_-_21630078 | 1.20 |
ENSMUST00000177577.2
|
1110059E24Rik
|
RIKEN cDNA 1110059E24 gene |
| chr17_+_24570991 | 1.17 |
ENSMUST00000039013.15
|
Abca3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr11_+_53661251 | 1.17 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr1_+_172309337 | 1.16 |
ENSMUST00000127052.8
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr1_-_155912216 | 1.14 |
ENSMUST00000027738.14
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr16_-_3726503 | 1.14 |
ENSMUST00000115859.8
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr9_+_81745723 | 1.14 |
ENSMUST00000057067.10
ENSMUST00000189832.7 ENSMUST00000189391.2 |
Mei4
|
meiotic double-stranded break formation protein 4 |
| chr15_-_100583044 | 1.13 |
ENSMUST00000230312.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr15_+_76555838 | 1.12 |
ENSMUST00000135388.3
|
Ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr12_-_69771604 | 1.12 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr16_+_31482658 | 1.11 |
ENSMUST00000115201.8
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr18_-_3281752 | 1.10 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr9_+_121245036 | 1.07 |
ENSMUST00000211187.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chrX_+_69429475 | 1.07 |
ENSMUST00000053981.6
|
Eola1
|
endothelium and lymphocyte associated ASCH domain 1 |
| chr12_+_105302853 | 1.05 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr17_+_24571011 | 1.04 |
ENSMUST00000079594.12
|
Abca3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr15_+_76555815 | 1.04 |
ENSMUST00000037551.15
|
Ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr17_+_28491085 | 1.04 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr9_-_121686643 | 1.04 |
ENSMUST00000215007.2
ENSMUST00000216914.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chrX_-_56549092 | 1.03 |
ENSMUST00000057645.6
|
Gpr101
|
G protein-coupled receptor 101 |
| chr8_+_91635192 | 1.02 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr6_-_23839419 | 0.99 |
ENSMUST00000115358.9
ENSMUST00000163871.9 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr6_-_33037107 | 0.99 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr17_-_24752683 | 0.99 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr8_-_84874468 | 0.99 |
ENSMUST00000117424.9
ENSMUST00000040383.9 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr7_-_41042684 | 0.98 |
ENSMUST00000071804.10
ENSMUST00000206801.2 ENSMUST00000205338.2 |
AI987944
|
expressed sequence AI987944 |
| chr5_+_38434738 | 0.98 |
ENSMUST00000114099.6
|
Otop1
|
otopetrin 1 |
| chr16_-_10131804 | 0.96 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2 |
| chr14_+_57661514 | 0.96 |
ENSMUST00000122063.8
|
Ift88
|
intraflagellar transport 88 |
| chr9_-_121686601 | 0.96 |
ENSMUST00000213124.2
ENSMUST00000215300.2 ENSMUST00000213147.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr4_+_95467653 | 0.95 |
ENSMUST00000043335.11
|
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr15_-_5137951 | 0.93 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr11_+_103540391 | 0.92 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chrX_-_165262631 | 0.92 |
ENSMUST00000049435.15
|
Rab9
|
RAB9, member RAS oncogene family |
| chr13_+_24118417 | 0.91 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr10_+_121477699 | 0.90 |
ENSMUST00000120642.9
ENSMUST00000132744.2 |
D930020B18Rik
|
RIKEN cDNA D930020B18 gene |
| chr14_+_31750946 | 0.90 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr4_+_40269563 | 0.90 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr18_-_7004717 | 0.87 |
ENSMUST00000079788.7
|
Mkx
|
mohawk homeobox |
| chr14_+_28233301 | 0.87 |
ENSMUST00000112272.2
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr3_+_80943667 | 0.85 |
ENSMUST00000029652.4
|
Pdgfc
|
platelet-derived growth factor, C polypeptide |
| chr1_+_185187000 | 0.85 |
ENSMUST00000061093.7
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr5_-_135807092 | 0.84 |
ENSMUST00000053906.11
ENSMUST00000177559.8 ENSMUST00000111161.9 ENSMUST00000111162.8 ENSMUST00000111163.9 |
Styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr15_-_5137975 | 0.84 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr7_-_118047394 | 0.84 |
ENSMUST00000203796.4
ENSMUST00000203485.3 |
Syt17
|
synaptotagmin XVII |
| chr15_+_39609320 | 0.83 |
ENSMUST00000227368.2
ENSMUST00000228556.2 ENSMUST00000022913.6 ENSMUST00000228701.2 ENSMUST00000227792.2 |
Dcstamp
|
dendrocyte expressed seven transmembrane protein |
| chr6_-_85742726 | 0.83 |
ENSMUST00000050780.8
|
Nat8f3
|
N-acetyltransferase 8 (GCN5-related) family member 3 |
| chr4_-_11386394 | 0.83 |
ENSMUST00000155519.2
|
Esrp1
|
epithelial splicing regulatory protein 1 |
| chrX_-_151552457 | 0.83 |
ENSMUST00000101141.9
ENSMUST00000062317.5 |
Shroom2
|
shroom family member 2 |
| chr9_+_110162470 | 0.82 |
ENSMUST00000198761.5
ENSMUST00000197630.3 |
Scap
|
SREBF chaperone |
| chr6_-_33037191 | 0.82 |
ENSMUST00000066379.11
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chrX_-_71318353 | 0.81 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr5_+_118307754 | 0.81 |
ENSMUST00000054836.7
|
Hrk
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr2_+_164327988 | 0.80 |
ENSMUST00000109350.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr3_+_87704258 | 0.79 |
ENSMUST00000029711.9
ENSMUST00000107582.3 |
Insrr
|
insulin receptor-related receptor |
| chr12_-_103623354 | 0.79 |
ENSMUST00000152517.2
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr14_+_61022006 | 0.78 |
ENSMUST00000063562.9
ENSMUST00000225043.2 |
Mipep
|
mitochondrial intermediate peptidase |
| chr16_+_31482745 | 0.78 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr17_+_34879431 | 0.77 |
ENSMUST00000238967.2
|
Tnxb
|
tenascin XB |
| chr5_-_30410720 | 0.77 |
ENSMUST00000088117.11
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr9_+_110709301 | 0.77 |
ENSMUST00000123389.8
|
Als2cl
|
ALS2 C-terminal like |
| chr17_-_66258110 | 0.77 |
ENSMUST00000233580.2
ENSMUST00000024906.6 |
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr3_-_108747767 | 0.77 |
ENSMUST00000196679.5
|
Stxbp3
|
syntaxin binding protein 3 |
| chr17_-_25564501 | 0.77 |
ENSMUST00000153118.2
ENSMUST00000146856.3 |
Tpsab1
|
tryptase alpha/beta 1 |
| chr3_-_10400710 | 0.76 |
ENSMUST00000078748.4
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr5_-_33786919 | 0.75 |
ENSMUST00000153696.8
ENSMUST00000045329.10 ENSMUST00000065119.15 |
Fam53a
|
family with sequence similarity 53, member A |
| chr9_+_65122381 | 0.74 |
ENSMUST00000213396.2
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr7_+_127440924 | 0.74 |
ENSMUST00000033075.14
|
Stx4a
|
syntaxin 4A (placental) |
| chr15_+_74828272 | 0.73 |
ENSMUST00000188042.2
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr12_-_17374704 | 0.72 |
ENSMUST00000020884.16
ENSMUST00000095820.12 ENSMUST00000221129.2 ENSMUST00000127185.8 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr19_-_10857734 | 0.72 |
ENSMUST00000133303.8
|
Tmem109
|
transmembrane protein 109 |
| chr6_+_107506678 | 0.71 |
ENSMUST00000049285.10
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chrX_-_100865583 | 0.71 |
ENSMUST00000239206.2
|
Gm3858
|
predicted gene 3858 |
| chr4_-_129083392 | 0.71 |
ENSMUST00000117497.8
ENSMUST00000117350.2 |
S100pbp
|
S100P binding protein |
| chr7_-_44465043 | 0.71 |
ENSMUST00000107893.9
|
Atf5
|
activating transcription factor 5 |
| chr17_-_26063391 | 0.71 |
ENSMUST00000176591.8
|
Rhot2
|
ras homolog family member T2 |
| chr4_-_129083439 | 0.70 |
ENSMUST00000106059.8
|
S100pbp
|
S100P binding protein |
| chr9_+_110709223 | 0.69 |
ENSMUST00000084926.9
|
Als2cl
|
ALS2 C-terminal like |
| chr9_+_110709353 | 0.68 |
ENSMUST00000155014.2
|
Als2cl
|
ALS2 C-terminal like |
| chr5_-_114411851 | 0.67 |
ENSMUST00000044790.12
|
Foxn4
|
forkhead box N4 |
| chr16_-_92262969 | 0.67 |
ENSMUST00000232239.2
ENSMUST00000060005.15 |
Rcan1
|
regulator of calcineurin 1 |
| chr14_+_119375753 | 0.67 |
ENSMUST00000065904.5
|
Hs6st3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr8_+_4399588 | 0.66 |
ENSMUST00000110982.8
ENSMUST00000024004.9 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr8_-_47986473 | 0.66 |
ENSMUST00000039061.15
|
Trappc11
|
trafficking protein particle complex 11 |
| chr4_-_143677586 | 0.66 |
ENSMUST00000105766.2
|
Pramel16
|
PRAME like 16 |
| chr9_+_110709260 | 0.65 |
ENSMUST00000130386.8
|
Als2cl
|
ALS2 C-terminal like |
| chr1_+_120268299 | 0.65 |
ENSMUST00000037286.12
|
C1ql2
|
complement component 1, q subcomponent-like 2 |
| chr1_+_192856044 | 0.64 |
ENSMUST00000193307.2
|
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Scrt1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 3.5 | 14.1 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 2.7 | 10.7 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.2 | 4.9 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 1.2 | 6.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 1.1 | 3.3 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 1.0 | 12.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 1.0 | 7.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.8 | 5.5 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.7 | 2.7 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.6 | 4.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.6 | 4.3 | GO:0060335 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) heme transport(GO:0015886) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.6 | 5.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.5 | 1.4 | GO:0007308 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.5 | 1.9 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.4 | 1.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.4 | 1.3 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.4 | 1.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.4 | 1.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.4 | 3.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.4 | 3.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 3.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 4.3 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.4 | 2.7 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.4 | 1.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.4 | 1.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 | 1.3 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.3 | 5.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 3.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 3.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 0.9 | GO:0060067 | chemorepulsion of dopaminergic neuron axon(GO:0036518) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.3 | 8.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 0.8 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.3 | 0.8 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.2 | 1.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 1.2 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.2 | 1.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 1.5 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 0.8 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.2 | 6.5 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.2 | 0.6 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.2 | 0.3 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.2 | 2.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 0.9 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.8 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 1.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.2 | 4.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.2 | 2.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.9 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.3 | GO:0042376 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 2.0 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 2.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.8 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.9 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 0.8 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 2.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.2 | GO:0060129 | corticotropin hormone secreting cell differentiation(GO:0060128) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 1.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 7.4 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 3.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 0.4 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.1 | 0.3 | GO:0071640 | macrophage inflammatory protein-1 alpha production(GO:0071608) regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.1 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 1.0 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 3.9 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.5 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 1.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.5 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 1.3 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.1 | 1.8 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.1 | 0.3 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 0.4 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 2.0 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.1 | 2.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.2 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 1.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.6 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 1.0 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.7 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.1 | 0.2 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.1 | 1.0 | GO:2001212 | renal system process involved in regulation of blood volume(GO:0001977) regulation of glomerular filtration(GO:0003093) caveola assembly(GO:0070836) regulation of vasculogenesis(GO:2001212) |
| 0.1 | 0.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 1.9 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 2.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 1.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 1.0 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 2.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 6.8 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.8 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.4 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.8 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 1.6 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 1.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 1.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.0 | GO:0035272 | exocrine system development(GO:0035272) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) regulation of neurotrophin production(GO:0032899) positive regulation of neurotrophin production(GO:0032901) |
| 0.0 | 0.1 | GO:0071043 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.8 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 1.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 5.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 7.2 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 2.3 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 4.4 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.6 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 2.2 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.2 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.0 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 19.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.8 | 7.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.7 | 2.2 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.4 | 4.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 1.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 1.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 1.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 3.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 0.7 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 3.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.0 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 1.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 11.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.9 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.1 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.5 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 1.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 10.7 | GO:0070160 | occluding junction(GO:0070160) |
| 0.0 | 9.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 6.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 5.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 1.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 47.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 2.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 11.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 5.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 24.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.5 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.2 | 4.9 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.2 | 3.6 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 1.2 | 9.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.1 | 5.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.9 | 4.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.8 | 3.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.8 | 2.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.7 | 2.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.7 | 4.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.7 | 16.8 | GO:0030228 | low-density lipoprotein particle binding(GO:0030169) lipoprotein particle receptor activity(GO:0030228) |
| 0.5 | 2.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.4 | 10.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 1.0 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.3 | 1.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 1.0 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.3 | 1.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 0.9 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.3 | 1.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 6.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 8.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 8.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.5 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 1.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 3.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 2.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 7.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 3.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 14.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 2.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.5 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.4 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.0 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.1 | 11.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.6 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 5.0 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 3.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.5 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 3.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 6.5 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 1.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.3 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 2.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 1.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 2.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 9.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 10.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 3.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 3.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 7.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 8.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 4.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 4.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 10.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 3.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 4.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 3.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 3.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 5.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |