Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
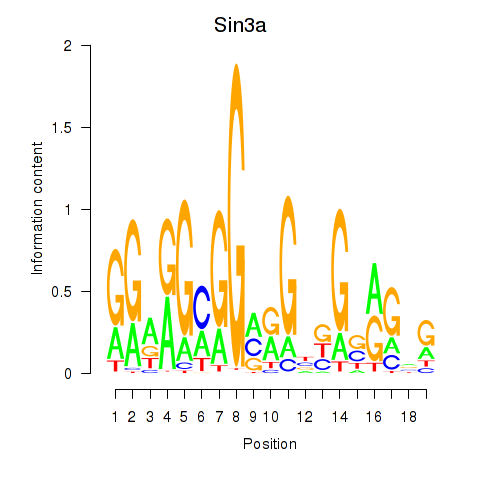
Results for Sin3a
Z-value: 0.92
Transcription factors associated with Sin3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sin3a
|
ENSMUSG00000042557.15 | transcriptional regulator, SIN3A (yeast) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sin3a | mm39_v1_chr9_+_56982622_56982727 | -0.65 | 2.0e-05 | Click! |
Activity profile of Sin3a motif
Sorted Z-values of Sin3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_46120327 | 5.68 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr2_+_155359868 | 1.89 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr2_+_155360015 | 1.73 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr19_-_4087940 | 1.70 |
ENSMUST00000237893.2
ENSMUST00000169613.4 |
Gstp1
|
glutathione S-transferase, pi 1 |
| chr16_-_67417768 | 1.61 |
ENSMUST00000114292.8
ENSMUST00000120898.8 |
Cadm2
|
cell adhesion molecule 2 |
| chr11_+_72326337 | 1.60 |
ENSMUST00000076443.10
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr16_-_67417687 | 1.55 |
ENSMUST00000120594.8
|
Cadm2
|
cell adhesion molecule 2 |
| chrX_-_146337046 | 1.45 |
ENSMUST00000112819.9
ENSMUST00000136789.8 |
Lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr7_+_16043502 | 1.40 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr2_+_58457370 | 1.36 |
ENSMUST00000071543.12
|
Upp2
|
uridine phosphorylase 2 |
| chr19_-_4087907 | 1.31 |
ENSMUST00000237982.2
|
Gstp1
|
glutathione S-transferase, pi 1 |
| chr17_-_28039506 | 1.31 |
ENSMUST00000114859.9
ENSMUST00000233533.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr11_+_72326391 | 1.25 |
ENSMUST00000100903.3
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr3_-_88857707 | 1.21 |
ENSMUST00000090938.11
|
Dap3
|
death associated protein 3 |
| chr19_-_4092218 | 1.20 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr14_+_70314652 | 1.19 |
ENSMUST00000035908.3
|
Egr3
|
early growth response 3 |
| chr2_+_78699360 | 1.19 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chrX_+_102400061 | 1.18 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr5_-_65593217 | 1.16 |
ENSMUST00000031103.14
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr7_-_126625657 | 1.15 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_+_72326358 | 1.15 |
ENSMUST00000108499.2
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chrX_-_103244728 | 1.13 |
ENSMUST00000056502.7
|
Nexmif
|
neurite extension and migration factor |
| chr7_+_116980173 | 1.10 |
ENSMUST00000032892.7
|
Xylt1
|
xylosyltransferase 1 |
| chr1_+_39232812 | 1.09 |
ENSMUST00000173050.8
|
Npas2
|
neuronal PAS domain protein 2 |
| chr15_-_66158445 | 1.08 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chr5_-_65585720 | 1.08 |
ENSMUST00000131263.3
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr6_-_83098255 | 1.08 |
ENSMUST00000205023.2
ENSMUST00000146328.4 ENSMUST00000151393.7 ENSMUST00000032111.11 ENSMUST00000113936.10 |
Wbp1
|
WW domain binding protein 1 |
| chr14_-_78970160 | 1.03 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr7_-_34353767 | 0.99 |
ENSMUST00000206501.2
ENSMUST00000108069.8 |
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr18_+_12776358 | 0.99 |
ENSMUST00000234966.2
ENSMUST00000025294.9 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr6_-_42301488 | 0.97 |
ENSMUST00000095974.4
|
Fam131b
|
family with sequence similarity 131, member B |
| chrX_-_103244784 | 0.97 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr13_+_31809774 | 0.96 |
ENSMUST00000042054.3
|
Foxf2
|
forkhead box F2 |
| chr7_-_126625739 | 0.95 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr14_-_24054186 | 0.95 |
ENSMUST00000188991.7
ENSMUST00000224468.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr4_+_107659361 | 0.95 |
ENSMUST00000106731.4
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr14_-_29443792 | 0.95 |
ENSMUST00000022567.9
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr7_+_131144596 | 0.95 |
ENSMUST00000046093.6
|
Hmx3
|
H6 homeobox 3 |
| chr5_+_122239007 | 0.94 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr5_+_122239030 | 0.92 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr2_-_52566583 | 0.91 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr7_-_24937276 | 0.91 |
ENSMUST00000071739.12
ENSMUST00000108411.2 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr17_-_28039588 | 0.90 |
ENSMUST00000114863.10
ENSMUST00000233131.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr2_+_102489558 | 0.90 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_-_5719302 | 0.89 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr2_+_102488985 | 0.89 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr6_+_99669640 | 0.89 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr9_-_107971729 | 0.88 |
ENSMUST00000193254.6
|
Apeh
|
acylpeptide hydrolase |
| chr15_-_3612628 | 0.87 |
ENSMUST00000110698.9
|
Ghr
|
growth hormone receptor |
| chr14_-_19057159 | 0.86 |
ENSMUST00000170123.2
|
Gm10409
|
predicted gene 10409 |
| chrX_-_103244703 | 0.86 |
ENSMUST00000087879.11
|
Nexmif
|
neurite extension and migration factor |
| chrX_+_68403900 | 0.85 |
ENSMUST00000033532.7
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr14_-_24054352 | 0.85 |
ENSMUST00000190339.2
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr4_+_107659474 | 0.84 |
ENSMUST00000106733.10
ENSMUST00000238651.2 ENSMUST00000030356.10 ENSMUST00000238421.2 ENSMUST00000126573.8 ENSMUST00000238569.2 ENSMUST00000106732.10 |
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr16_-_20440005 | 0.84 |
ENSMUST00000052939.4
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr1_-_57010921 | 0.84 |
ENSMUST00000114415.10
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr4_-_129142208 | 0.83 |
ENSMUST00000052602.6
|
C77080
|
expressed sequence C77080 |
| chr6_-_48422447 | 0.83 |
ENSMUST00000114564.8
|
Zfp467
|
zinc finger protein 467 |
| chr1_+_60448931 | 0.83 |
ENSMUST00000189082.7
ENSMUST00000187709.7 |
Abi2
|
abl interactor 2 |
| chr17_+_26332260 | 0.83 |
ENSMUST00000235821.2
ENSMUST00000025010.14 ENSMUST00000237058.2 |
Pgap6
|
post-glycosylphosphatidylinositol attachment to proteins 6 |
| chr1_+_133291302 | 0.81 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr14_-_24054273 | 0.81 |
ENSMUST00000188285.7
ENSMUST00000190044.7 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_+_27567213 | 0.80 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr3_-_89230190 | 0.80 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr4_+_119396913 | 0.80 |
ENSMUST00000137560.8
|
Foxj3
|
forkhead box J3 |
| chr3_-_88857578 | 0.79 |
ENSMUST00000174402.8
ENSMUST00000174077.8 |
Dap3
|
death associated protein 3 |
| chr9_-_106769131 | 0.78 |
ENSMUST00000159283.8
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr11_+_69826719 | 0.78 |
ENSMUST00000149194.8
|
Ybx2
|
Y box protein 2 |
| chrX_+_100298134 | 0.78 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr5_+_36641922 | 0.78 |
ENSMUST00000060100.3
|
Ccdc96
|
coiled-coil domain containing 96 |
| chr8_+_106052970 | 0.78 |
ENSMUST00000015000.12
ENSMUST00000098453.9 |
Tmem208
|
transmembrane protein 208 |
| chr7_+_45349267 | 0.77 |
ENSMUST00000003360.10
|
Car11
|
carbonic anhydrase 11 |
| chrX_-_59937036 | 0.77 |
ENSMUST00000135107.4
|
Sox3
|
SRY (sex determining region Y)-box 3 |
| chr14_-_19635203 | 0.77 |
ENSMUST00000170694.9
|
Gm2237
|
predicted gene 2237 |
| chr19_-_46136765 | 0.77 |
ENSMUST00000026259.16
|
Pitx3
|
paired-like homeodomain transcription factor 3 |
| chr15_+_87509413 | 0.77 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr7_+_123582021 | 0.76 |
ENSMUST00000106437.2
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr7_-_117842892 | 0.76 |
ENSMUST00000179047.3
|
Smg1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
| chr4_-_57143437 | 0.76 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr7_-_74204474 | 0.75 |
ENSMUST00000107453.8
|
Slco3a1
|
solute carrier organic anion transporter family, member 3a1 |
| chr18_+_69479211 | 0.75 |
ENSMUST00000201235.4
|
Tcf4
|
transcription factor 4 |
| chr17_-_74601769 | 0.75 |
ENSMUST00000078459.8
ENSMUST00000232989.2 |
Memo1
|
mediator of cell motility 1 |
| chr13_+_8935537 | 0.74 |
ENSMUST00000169314.9
|
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr2_+_139520098 | 0.73 |
ENSMUST00000184404.8
ENSMUST00000099307.4 |
Ism1
|
isthmin 1, angiogenesis inhibitor |
| chr11_+_69826603 | 0.73 |
ENSMUST00000018698.12
|
Ybx2
|
Y box protein 2 |
| chr17_-_74601828 | 0.72 |
ENSMUST00000233514.3
|
Memo1
|
mediator of cell motility 1 |
| chr19_-_43663282 | 0.72 |
ENSMUST00000046038.9
ENSMUST00000236433.2 |
Slc25a28
|
solute carrier family 25, member 28 |
| chr7_-_119494669 | 0.72 |
ENSMUST00000098080.9
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr2_-_25359752 | 0.72 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr9_-_106769069 | 0.71 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr19_+_53891730 | 0.71 |
ENSMUST00000025931.14
|
Pdcd4
|
programmed cell death 4 |
| chr14_-_18287197 | 0.71 |
ENSMUST00000164512.8
|
Gm2974
|
predicted gene 2974 |
| chr7_+_36397426 | 0.71 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr1_-_65225617 | 0.70 |
ENSMUST00000186222.7
ENSMUST00000169032.8 ENSMUST00000191459.2 ENSMUST00000188876.7 |
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr9_-_78254422 | 0.70 |
ENSMUST00000034902.12
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr7_+_100970435 | 0.70 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr1_+_167426019 | 0.69 |
ENSMUST00000111386.8
ENSMUST00000111384.8 |
Rxrg
|
retinoid X receptor gamma |
| chr10_-_43050516 | 0.69 |
ENSMUST00000040275.9
|
Sobp
|
sine oculis binding protein |
| chr1_+_171157137 | 0.69 |
ENSMUST00000142063.8
ENSMUST00000129116.8 |
Dedd
|
death effector domain-containing |
| chr16_-_18245352 | 0.69 |
ENSMUST00000000335.12
|
Comt
|
catechol-O-methyltransferase |
| chr14_-_17742998 | 0.69 |
ENSMUST00000165619.8
|
Gm3252
|
predicted gene 3252 |
| chr7_+_44078366 | 0.68 |
ENSMUST00000127790.8
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr9_-_78254443 | 0.68 |
ENSMUST00000129247.2
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr11_+_78215026 | 0.68 |
ENSMUST00000102478.4
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr11_+_69871952 | 0.68 |
ENSMUST00000108593.8
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr1_+_182591771 | 0.68 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr1_-_75119277 | 0.67 |
ENSMUST00000168720.8
ENSMUST00000041213.12 ENSMUST00000189809.2 |
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr4_-_91260265 | 0.67 |
ENSMUST00000107110.8
ENSMUST00000008633.15 ENSMUST00000107118.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr1_+_166081664 | 0.67 |
ENSMUST00000111416.7
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr12_+_110413523 | 0.67 |
ENSMUST00000222276.2
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_66888474 | 0.67 |
ENSMUST00000162439.8
ENSMUST00000162098.9 |
Shox2
|
short stature homeobox 2 |
| chr14_-_16968099 | 0.67 |
ENSMUST00000181562.8
|
Gm3488
|
predicted gene, 3488 |
| chrX_+_72760183 | 0.66 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr19_-_6134903 | 0.66 |
ENSMUST00000160977.8
ENSMUST00000159859.2 ENSMUST00000025707.9 ENSMUST00000160712.8 ENSMUST00000237738.2 |
Zfpl1
|
zinc finger like protein 1 |
| chr10_-_84938350 | 0.66 |
ENSMUST00000059383.8
ENSMUST00000216889.2 |
Fhl4
|
four and a half LIM domains 4 |
| chr5_-_33591320 | 0.66 |
ENSMUST00000173348.2
|
Nkx1-1
|
NK1 homeobox 1 |
| chr14_+_70314727 | 0.66 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr17_+_9020522 | 0.66 |
ENSMUST00000089085.10
|
Pde10a
|
phosphodiesterase 10A |
| chr5_+_91175323 | 0.65 |
ENSMUST00000202724.4
ENSMUST00000041516.9 |
Epgn
|
epithelial mitogen |
| chr2_+_152873772 | 0.65 |
ENSMUST00000037235.7
|
Xkr7
|
X-linked Kx blood group related 7 |
| chr7_+_119495058 | 0.65 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr7_-_132724344 | 0.65 |
ENSMUST00000167218.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr14_-_49763310 | 0.65 |
ENSMUST00000146164.2
ENSMUST00000138884.8 ENSMUST00000074368.11 ENSMUST00000123534.2 |
Slc35f4
|
solute carrier family 35, member F4 |
| chr1_+_42992109 | 0.65 |
ENSMUST00000179766.3
|
Gpr45
|
G protein-coupled receptor 45 |
| chr14_-_18659699 | 0.65 |
ENSMUST00000170480.8
|
Gm3002
|
predicted gene 3002 |
| chr14_+_15579811 | 0.64 |
ENSMUST00000171906.2
|
Gm3667
|
predicted gene 3667 |
| chr15_-_76406102 | 0.64 |
ENSMUST00000164703.2
|
Scrt1
|
scratch family zinc finger 1 |
| chr14_-_18817743 | 0.64 |
ENSMUST00000167430.8
|
Gm3020
|
predicted gene 3020 |
| chr12_-_5425682 | 0.63 |
ENSMUST00000020958.9
|
Klhl29
|
kelch-like 29 |
| chr19_+_6547790 | 0.63 |
ENSMUST00000113458.8
ENSMUST00000113459.2 |
Nrxn2
|
neurexin II |
| chrX_+_57076359 | 0.63 |
ENSMUST00000088631.11
ENSMUST00000088629.4 |
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr10_-_80235384 | 0.63 |
ENSMUST00000092295.10
ENSMUST00000105349.8 |
Mbd3
|
methyl-CpG binding domain protein 3 |
| chr2_-_84605732 | 0.62 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr11_-_115078653 | 0.62 |
ENSMUST00000103041.8
|
Nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr2_-_25360043 | 0.62 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr6_-_116170389 | 0.62 |
ENSMUST00000088896.10
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr7_-_43139390 | 0.62 |
ENSMUST00000107974.3
|
Iglon5
|
IgLON family member 5 |
| chr17_+_85928459 | 0.62 |
ENSMUST00000162695.3
|
Six3
|
sine oculis-related homeobox 3 |
| chr7_+_57240894 | 0.62 |
ENSMUST00000039697.14
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chrX_-_161426542 | 0.62 |
ENSMUST00000101102.2
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chrX_-_20483845 | 0.61 |
ENSMUST00000116621.2
|
Ndufb11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr9_-_86762450 | 0.61 |
ENSMUST00000191290.3
|
Snap91
|
synaptosomal-associated protein 91 |
| chr11_-_4696778 | 0.61 |
ENSMUST00000009219.3
|
Cabp7
|
calcium binding protein 7 |
| chr7_+_27291126 | 0.60 |
ENSMUST00000167435.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr6_+_125298372 | 0.60 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chrX_+_5959507 | 0.60 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr7_-_132725041 | 0.60 |
ENSMUST00000171022.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr1_+_167425953 | 0.60 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma |
| chr19_-_6134703 | 0.60 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1 |
| chr4_-_3938352 | 0.60 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr7_-_119494918 | 0.60 |
ENSMUST00000059851.14
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr14_-_24053994 | 0.60 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_-_6085069 | 0.60 |
ENSMUST00000103006.4
|
Nudt10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr12_+_21161722 | 0.59 |
ENSMUST00000064595.15
ENSMUST00000101562.11 ENSMUST00000090834.13 |
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr15_+_26309125 | 0.59 |
ENSMUST00000126304.2
ENSMUST00000140840.8 ENSMUST00000152841.2 |
Marchf11
|
membrane associated ring-CH-type finger 11 |
| chr1_-_75294234 | 0.59 |
ENSMUST00000066668.14
ENSMUST00000185797.7 |
Dnpep
|
aspartyl aminopeptidase |
| chr3_-_88997261 | 0.59 |
ENSMUST00000196223.5
ENSMUST00000196043.2 ENSMUST00000166687.6 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr4_+_134124691 | 0.59 |
ENSMUST00000105870.8
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr8_+_120955195 | 0.58 |
ENSMUST00000180448.3
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr4_-_104967032 | 0.58 |
ENSMUST00000030243.8
|
Prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr9_-_107971640 | 0.58 |
ENSMUST00000081309.13
ENSMUST00000191985.2 |
Apeh
|
acylpeptide hydrolase |
| chr19_+_53891802 | 0.58 |
ENSMUST00000165617.3
|
Pdcd4
|
programmed cell death 4 |
| chr10_+_29023201 | 0.58 |
ENSMUST00000213243.2
|
Soga3
|
SOGA family member 3 |
| chr7_-_34354924 | 0.58 |
ENSMUST00000032709.3
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr11_-_72686853 | 0.58 |
ENSMUST00000156294.8
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr6_-_42301574 | 0.57 |
ENSMUST00000031891.15
ENSMUST00000143278.8 |
Fam131b
|
family with sequence similarity 131, member B |
| chr7_+_19016536 | 0.57 |
ENSMUST00000032559.17
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr19_+_53665719 | 0.57 |
ENSMUST00000164202.9
|
Rbm20
|
RNA binding motif protein 20 |
| chr14_+_122712809 | 0.57 |
ENSMUST00000075888.6
|
Zic2
|
zinc finger protein of the cerebellum 2 |
| chr11_+_45871135 | 0.57 |
ENSMUST00000049038.4
|
Sox30
|
SRY (sex determining region Y)-box 30 |
| chr6_-_52237765 | 0.57 |
ENSMUST00000147595.7
|
Hoxa13
|
homeobox A13 |
| chr15_-_3612703 | 0.56 |
ENSMUST00000069451.11
|
Ghr
|
growth hormone receptor |
| chr14_-_18897750 | 0.56 |
ENSMUST00000178728.2
|
Gm3005
|
predicted gene 3005 |
| chr14_+_14901127 | 0.56 |
ENSMUST00000163790.2
|
Gm3558
|
predicted gene 3558 |
| chr10_-_31485180 | 0.56 |
ENSMUST00000081989.8
|
Rnf217
|
ring finger protein 217 |
| chr11_-_78313043 | 0.56 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr10_+_39608094 | 0.56 |
ENSMUST00000019986.13
|
Rev3l
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr8_+_87350672 | 0.56 |
ENSMUST00000034141.18
ENSMUST00000122188.10 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr7_+_112806672 | 0.56 |
ENSMUST00000047321.9
ENSMUST00000210074.2 ENSMUST00000210238.2 |
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr12_-_65012270 | 0.56 |
ENSMUST00000222508.2
|
Klhl28
|
kelch-like 28 |
| chr7_+_3439144 | 0.56 |
ENSMUST00000182222.8
|
Cacng8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr11_-_68277799 | 0.56 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chr11_-_88608958 | 0.56 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_+_126808016 | 0.56 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr1_+_75119419 | 0.55 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr16_+_91022300 | 0.55 |
ENSMUST00000035608.10
|
Olig2
|
oligodendrocyte transcription factor 2 |
| chr11_+_70861007 | 0.55 |
ENSMUST00000018593.10
|
Rpain
|
RPA interacting protein |
| chr7_-_126625617 | 0.55 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr1_+_171156942 | 0.55 |
ENSMUST00000111299.8
ENSMUST00000064950.11 |
Dedd
|
death effector domain-containing |
| chrX_+_59591614 | 0.55 |
ENSMUST00000117865.3
|
Gm715
|
predicted gene 715 |
| chr5_-_23881353 | 0.54 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr14_-_19420488 | 0.54 |
ENSMUST00000166494.2
|
Gm2897
|
predicted gene 2897 |
| chr4_+_140941259 | 0.54 |
ENSMUST00000097813.3
|
Cplane2
|
ciliogenesis and planar polarity effector 2 |
| chr5_-_46014809 | 0.54 |
ENSMUST00000190036.7
ENSMUST00000189859.7 ENSMUST00000186633.3 ENSMUST00000016026.14 ENSMUST00000045586.13 ENSMUST00000238522.2 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr7_+_24310738 | 0.54 |
ENSMUST00000073325.6
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr6_-_72876882 | 0.54 |
ENSMUST00000068697.11
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr10_-_75353157 | 0.54 |
ENSMUST00000039796.14
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr12_-_84745388 | 0.53 |
ENSMUST00000221905.2
ENSMUST00000222422.2 ENSMUST00000095550.4 |
Syndig1l
|
synapse differentiation inducing 1 like |
| chr17_+_31605184 | 0.53 |
ENSMUST00000047168.13
ENSMUST00000127929.8 ENSMUST00000134525.9 ENSMUST00000236454.2 ENSMUST00000238091.2 ENSMUST00000235719.2 |
Pde9a
|
phosphodiesterase 9A |
| chr1_-_37903881 | 0.53 |
ENSMUST00000239367.2
ENSMUST00000123082.9 |
Tsga10
|
testis specific 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sin3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 1.0 | 3.0 | GO:2000469 | negative regulation of peroxidase activity(GO:2000469) |
| 0.6 | 1.9 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.5 | 3.2 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.5 | 1.5 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.5 | 5.7 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 1.3 | GO:0060939 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.4 | 1.3 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.3 | 1.0 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.3 | 1.7 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.3 | 1.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 0.9 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.3 | 1.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.3 | 4.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 1.4 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.3 | 3.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 0.8 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.3 | 0.8 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.3 | 0.5 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.3 | 1.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 1.6 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 0.8 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.3 | 1.8 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 0.7 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.2 | 1.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 0.7 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 0.2 | 2.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.2 | 0.7 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.2 | 1.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.2 | 0.4 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 0.6 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 1.0 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.2 | 0.6 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.2 | 0.8 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 0.6 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.2 | 0.4 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 0.6 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.1 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.2 | 2.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.2 | 0.9 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.2 | 0.7 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.2 | 0.9 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 0.5 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.2 | 1.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 | 1.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 1.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 0.2 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.2 | 1.7 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 0.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 1.0 | GO:0021623 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.2 | 1.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 0.5 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 0.5 | GO:0018900 | dichloromethane metabolic process(GO:0018900) |
| 0.2 | 0.5 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.2 | 0.3 | GO:1900238 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.2 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.4 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.1 | 1.0 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.4 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.1 | 1.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.6 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.4 | GO:0003167 | atrioventricular bundle cell differentiation(GO:0003167) |
| 0.1 | 0.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 1.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.2 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 2.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.5 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.7 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.5 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 1.6 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.7 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.6 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.0 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.3 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 0.3 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.1 | 0.5 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.1 | 0.6 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.3 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 1.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.8 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 1.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.3 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.1 | 0.3 | GO:2000612 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.3 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.7 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 0.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.5 | GO:0060578 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.6 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.4 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.5 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.5 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.3 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.3 | GO:0090427 | activation of meiosis involved in egg activation(GO:0060466) activation of meiosis(GO:0090427) |
| 0.1 | 0.3 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.2 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.3 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.3 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.3 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.3 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 1.0 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.5 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.3 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.2 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.2 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.7 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.2 | GO:0061193 | formation of anatomical boundary(GO:0048859) taste bud development(GO:0061193) |
| 0.1 | 0.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.7 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.5 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 0.5 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.3 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.8 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.4 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.5 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.2 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.4 | GO:0072240 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 0.6 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.3 | GO:0021941 | radial glia guided migration of cerebellar granule cell(GO:0021933) negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.1 | 0.6 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.6 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 1.5 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.5 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 0.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.5 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 1.2 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.5 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.3 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 0.3 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.4 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.8 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.1 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.1 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 1.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.6 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.1 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.2 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.4 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.1 | 0.2 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.2 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.4 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.9 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.3 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.2 | GO:1904456 | negative regulation of neuronal action potential(GO:1904456) |
| 0.1 | 0.9 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.8 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.2 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 0.2 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.2 | GO:1901254 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 1.2 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.6 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.3 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 1.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.5 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 0.5 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.2 | GO:1990036 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.1 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 1.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.4 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.1 | 0.4 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 1.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.5 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0046666 | retinal cell programmed cell death(GO:0046666) |
| 0.0 | 1.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.2 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.3 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.2 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.3 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.2 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.9 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 2.9 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.4 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 1.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.6 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.0 | 1.5 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.2 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0019452 | L-cysteine catabolic process to taurine(GO:0019452) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.6 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.3 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.6 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.8 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.0 | 0.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.5 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.2 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.1 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.2 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.4 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.4 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.0 | 0.5 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.6 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:0050822 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.5 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.4 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.1 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.7 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.3 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.0 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0071372 | response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.6 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.2 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.2 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0070375 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) ERK5 cascade(GO:0070375) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 1.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.3 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.8 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.4 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.5 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 1.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.6 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.4 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.0 | GO:2000617 | positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.4 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.3 | GO:0030826 | regulation of cGMP biosynthetic process(GO:0030826) |
| 0.0 | 0.2 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) |
| 0.0 | 0.5 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.0 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 0.1 | GO:0007296 | pericardium morphogenesis(GO:0003344) vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:1903935 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.5 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.6 | 1.9 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 1.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 1.0 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.3 | 1.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 1.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 3.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 0.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 0.6 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 0.7 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.2 | 0.7 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 0.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 1.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 2.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.2 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.1 | 0.5 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.1 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 2.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 3.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.2 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 1.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 2.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 8.6 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0060473 | cortical granule(GO:0060473) |
| 0.0 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.5 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 2.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.4 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.0 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.3 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.0 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.0 | 3.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.7 | 3.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.6 | 1.9 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.5 | 5.7 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 3.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 1.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.4 | 1.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.3 | 1.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 4.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 1.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.3 | 2.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 1.2 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 0.7 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 0.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 1.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 0.8 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.2 | 0.6 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.2 | 0.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.2 | 0.8 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.2 | 0.8 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.2 | 0.9 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 1.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 0.5 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.2 | 0.5 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 0.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 0.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 0.6 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 0.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 0.5 | GO:0016824 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.1 | 1.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.0 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.0 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.1 | 0.4 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 0.4 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.4 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.8 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.4 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.1 | 0.8 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 1.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.9 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.4 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.1 | 1.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.4 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.2 | GO:0034012 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 4.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.3 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 1.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.3 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 3.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.3 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.1 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.0 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 3.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.4 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.3 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 1.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.4 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 1.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0038100 | nodal binding(GO:0038100) |
| 0.0 | 1.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0001571 | non-tyrosine kinase fibroblast growth factor receptor activity(GO:0001571) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.1 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.1 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.0 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.5 | GO:0046977 | TAP binding(GO:0046977) |
| 0.0 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 1.0 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.8 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 1.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.4 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.4 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 8.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.5 | GO:0008066 | ionotropic glutamate receptor activity(GO:0004970) extracellular-glutamate-gated ion channel activity(GO:0005234) glutamate receptor activity(GO:0008066) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 0.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 2.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 3.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 7.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 4.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 3.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |