Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Six3_Six1_Six2
Z-value: 0.57
Transcription factors associated with Six3_Six1_Six2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
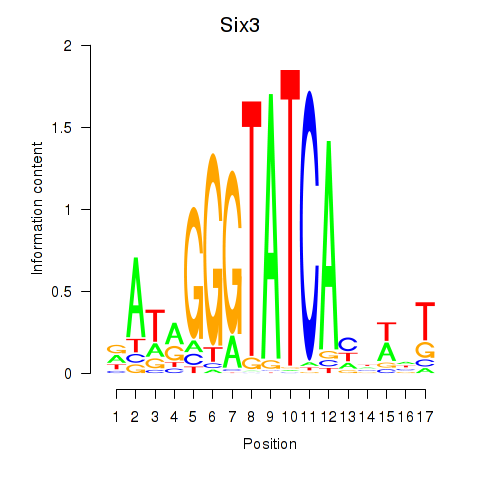
Six3
|
ENSMUSG00000038805.11 | sine oculis-related homeobox 3 |
|
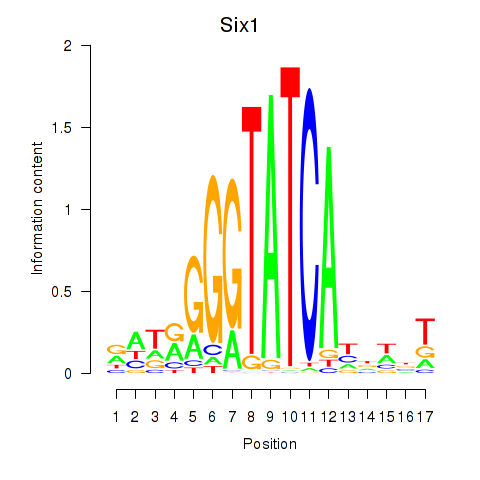
Six1
|
ENSMUSG00000051367.9 | sine oculis-related homeobox 1 |
|
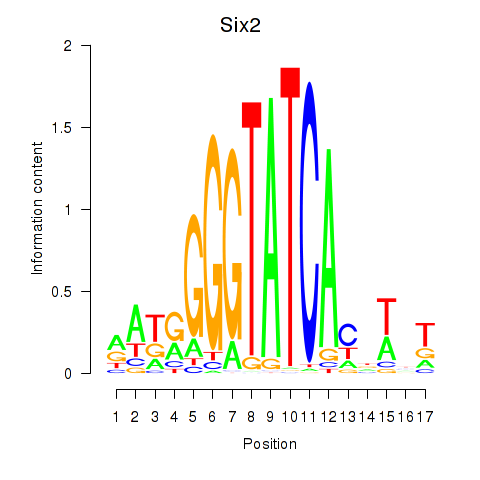
Six2
|
ENSMUSG00000024134.12 | sine oculis-related homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six2 | mm39_v1_chr17_-_85995680_85995703 | 0.25 | 1.3e-01 | Click! |
| Six3 | mm39_v1_chr17_+_85928459_85928469 | 0.23 | 1.8e-01 | Click! |
| Six1 | mm39_v1_chr12_-_73093953_73093953 | 0.19 | 2.6e-01 | Click! |
Activity profile of Six3_Six1_Six2 motif
Sorted Z-values of Six3_Six1_Six2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_117688486 | 14.38 |
ENSMUST00000106331.2
|
6030468B19Rik
|
RIKEN cDNA 6030468B19 gene |
| chr9_+_110856425 | 5.19 |
ENSMUST00000199313.2
|
Ltf
|
lactotransferrin |
| chr3_-_37778470 | 3.50 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr17_+_41121979 | 3.08 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr7_+_89814713 | 2.54 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_-_141749704 | 2.46 |
ENSMUST00000041317.3
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr6_+_123099615 | 2.41 |
ENSMUST00000161636.8
ENSMUST00000161365.8 |
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr14_+_56003406 | 2.34 |
ENSMUST00000057569.4
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr17_+_35460722 | 2.20 |
ENSMUST00000068056.12
ENSMUST00000174757.8 ENSMUST00000173731.8 |
Ddx39b
|
DEAD box helicase 39b |
| chr6_+_123100382 | 1.78 |
ENSMUST00000032248.8
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr6_+_123100272 | 1.70 |
ENSMUST00000041779.13
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr1_+_139382485 | 1.70 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr9_+_98305014 | 1.53 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chr15_-_82128888 | 1.41 |
ENSMUST00000089155.6
ENSMUST00000089157.11 |
Cenpm
|
centromere protein M |
| chr15_-_103218876 | 1.22 |
ENSMUST00000079824.6
|
Gpr84
|
G protein-coupled receptor 84 |
| chrX_-_73416824 | 1.22 |
ENSMUST00000178691.2
ENSMUST00000114146.8 |
Ubl4a
Slc10a3
|
ubiquitin-like 4A solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr19_-_38032006 | 1.20 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr3_+_32760447 | 1.19 |
ENSMUST00000194781.6
|
Actl6a
|
actin-like 6A |
| chrX_-_73416869 | 1.11 |
ENSMUST00000073067.11
ENSMUST00000037967.6 |
Slc10a3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr1_+_136395673 | 1.06 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr10_-_81463631 | 1.05 |
ENSMUST00000042923.9
|
Sirt6
|
sirtuin 6 |
| chr8_+_95720864 | 1.04 |
ENSMUST00000212141.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr2_+_127178072 | 0.98 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr7_-_79765042 | 0.95 |
ENSMUST00000206714.2
ENSMUST00000107384.10 |
Idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr7_-_100504610 | 0.92 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr2_-_88534814 | 0.87 |
ENSMUST00000216928.2
ENSMUST00000216977.2 |
Olfr1196
|
olfactory receptor 1196 |
| chr12_-_114252202 | 0.87 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr19_-_38031774 | 0.84 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr2_+_31135813 | 0.70 |
ENSMUST00000000199.8
|
Ncs1
|
neuronal calcium sensor 1 |
| chr13_+_104365880 | 0.67 |
ENSMUST00000022227.8
|
Cenpk
|
centromere protein K |
| chr5_+_140593075 | 0.66 |
ENSMUST00000031555.3
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_+_79048884 | 0.62 |
ENSMUST00000137667.3
|
Fanci
|
Fanconi anemia, complementation group I |
| chr15_+_101152078 | 0.56 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr17_-_53846438 | 0.55 |
ENSMUST00000056198.4
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr8_+_57908920 | 0.54 |
ENSMUST00000034023.4
|
Scrg1
|
scrapie responsive gene 1 |
| chr17_+_36176485 | 0.52 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr5_+_21748523 | 0.51 |
ENSMUST00000035651.6
|
Lrrc17
|
leucine rich repeat containing 17 |
| chr15_+_82136598 | 0.51 |
ENSMUST00000136948.3
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr12_-_113589576 | 0.49 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chrX_+_168468186 | 0.48 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr6_-_119925387 | 0.48 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr2_-_13276074 | 0.46 |
ENSMUST00000137670.3
ENSMUST00000114791.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr2_-_13276205 | 0.45 |
ENSMUST00000191959.6
ENSMUST00000028059.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr8_-_4325886 | 0.44 |
ENSMUST00000003029.14
|
Timm44
|
translocase of inner mitochondrial membrane 44 |
| chr2_+_106523532 | 0.41 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr6_-_120893771 | 0.41 |
ENSMUST00000004560.12
|
Bid
|
BH3 interacting domain death agonist |
| chr19_+_46329552 | 0.39 |
ENSMUST00000128041.8
|
Mfsd13a
|
major facilitator superfamily domain containing 13a |
| chr2_-_118859821 | 0.38 |
ENSMUST00000110833.2
ENSMUST00000036470.14 ENSMUST00000110834.8 |
Ccdc32
|
coiled-coil domain containing 32 |
| chr8_+_70285282 | 0.38 |
ENSMUST00000131637.9
|
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr7_-_101552989 | 0.37 |
ENSMUST00000106969.9
|
Tomt
|
transmembrane O-methyltransferase |
| chr7_-_104991477 | 0.37 |
ENSMUST00000213290.2
|
Olfr691
|
olfactory receptor 691 |
| chr11_-_99121822 | 0.34 |
ENSMUST00000103133.4
|
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr8_+_85558151 | 0.34 |
ENSMUST00000036734.6
|
Gadd45gip1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr2_+_117080212 | 0.34 |
ENSMUST00000028825.5
|
Fam98b
|
family with sequence similarity 98, member B |
| chr4_+_132903646 | 0.31 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr4_+_11758147 | 0.30 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr9_-_119170456 | 0.29 |
ENSMUST00000139870.2
|
Myd88
|
myeloid differentiation primary response gene 88 |
| chr2_+_86128161 | 0.26 |
ENSMUST00000054746.5
|
Olfr1052
|
olfactory receptor 1052 |
| chr17_-_35978438 | 0.26 |
ENSMUST00000043674.15
|
Vars2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr8_+_70285133 | 0.25 |
ENSMUST00000081503.13
|
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr7_-_103778992 | 0.25 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr19_-_5452521 | 0.24 |
ENSMUST00000235569.2
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr6_-_69741999 | 0.23 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr6_-_57827328 | 0.22 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr9_+_106099797 | 0.22 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr14_+_58035640 | 0.22 |
ENSMUST00000111269.2
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr9_+_38725910 | 0.22 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922 |
| chr1_-_65162267 | 0.22 |
ENSMUST00000050047.4
ENSMUST00000148020.8 |
D630023F18Rik
|
RIKEN cDNA D630023F18 gene |
| chr18_+_37840092 | 0.21 |
ENSMUST00000195823.2
|
Pcdhga6
|
protocadherin gamma subfamily A, 6 |
| chr2_-_89408791 | 0.20 |
ENSMUST00000217402.2
|
Olfr1245
|
olfactory receptor 1245 |
| chr16_-_18052937 | 0.19 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr9_+_65048454 | 0.18 |
ENSMUST00000034961.6
ENSMUST00000217371.2 |
Igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr10_-_128016135 | 0.14 |
ENSMUST00000238843.2
ENSMUST00000099139.9 |
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr8_-_87611849 | 0.14 |
ENSMUST00000034074.8
|
N4bp1
|
NEDD4 binding protein 1 |
| chr11_+_101877876 | 0.11 |
ENSMUST00000010985.8
|
Cfap97d1
|
CFAP97 domain containing 1 |
| chr12_-_75596441 | 0.11 |
ENSMUST00000218716.2
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr14_+_51181956 | 0.11 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr18_+_89224219 | 0.09 |
ENSMUST00000236835.2
|
Cd226
|
CD226 antigen |
| chr6_-_57306479 | 0.09 |
ENSMUST00000227283.2
ENSMUST00000228356.2 |
Vmn1r16
|
vomeronasal 1 receptor 16 |
| chr14_+_54440591 | 0.07 |
ENSMUST00000103725.2
|
Traj16
|
T cell receptor alpha joining 16 |
| chr19_+_9824919 | 0.07 |
ENSMUST00000179814.3
|
Scgb2a2
|
secretoglobin, family 2A, member 2 |
| chr2_+_157870399 | 0.06 |
ENSMUST00000103123.10
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr17_+_47221377 | 0.04 |
ENSMUST00000024773.6
|
Prph2
|
peripherin 2 |
| chr1_+_40305738 | 0.04 |
ENSMUST00000114795.3
|
Il1r1
|
interleukin 1 receptor, type I |
| chr7_-_102507962 | 0.03 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr11_+_49379915 | 0.03 |
ENSMUST00000214948.2
|
Olfr1385
|
olfactory receptor 1385 |
| chr13_+_94219934 | 0.02 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr6_-_120893725 | 0.02 |
ENSMUST00000145948.2
|
Bid
|
BH3 interacting domain death agonist |
| chr4_-_32950812 | 0.01 |
ENSMUST00000084750.8
ENSMUST00000084748.9 |
Ankrd6
|
ankyrin repeat domain 6 |
| chr6_+_90246088 | 0.01 |
ENSMUST00000058039.3
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr9_+_38686470 | 0.01 |
ENSMUST00000071681.4
|
Olfr921
|
olfactory receptor 921 |
| chr12_-_114451189 | 0.01 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six3_Six1_Six2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0051673 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.8 | 3.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.5 | 1.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.4 | 2.5 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.4 | 1.1 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 0.3 | 1.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 0.9 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.3 | 2.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.3 | 1.7 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.7 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 2.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.6 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.2 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.1 | 0.3 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 0.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.4 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.4 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 1.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 2.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0060369 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.9 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 7.2 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.0 | GO:0010286 | heat acclimation(GO:0010286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.2 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 2.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 2.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 1.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 1.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.9 | GO:0005901 | caveola(GO:0005901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 2.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 1.0 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.3 | 2.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.9 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 2.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 5.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 3.1 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 5.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |