Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
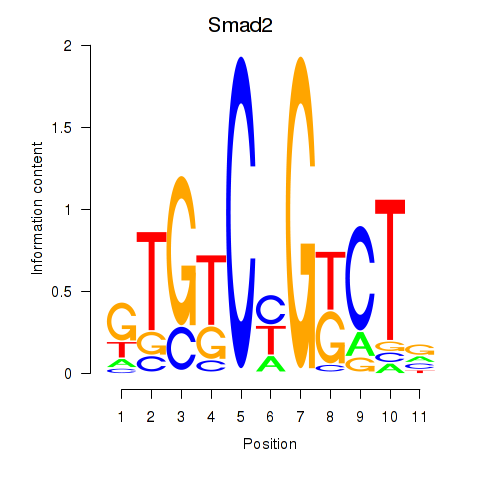
Results for Smad2
Z-value: 0.41
Transcription factors associated with Smad2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad2
|
ENSMUSG00000024563.17 | SMAD family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad2 | mm39_v1_chr18_+_76375206_76375245 | 0.37 | 2.5e-02 | Click! |
Activity profile of Smad2 motif
Sorted Z-values of Smad2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_30639217 | 4.63 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr7_+_19144950 | 2.61 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr7_-_126062272 | 2.37 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr15_-_101833160 | 0.75 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr1_-_172125555 | 0.66 |
ENSMUST00000085913.11
ENSMUST00000097464.4 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr2_-_148285450 | 0.56 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr3_-_116762617 | 0.50 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr2_+_38229270 | 0.42 |
ENSMUST00000143783.9
|
Lhx2
|
LIM homeobox protein 2 |
| chr5_-_113938417 | 0.38 |
ENSMUST00000160374.2
ENSMUST00000067853.6 |
Tmem119
|
transmembrane protein 119 |
| chr2_+_173918715 | 0.35 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr7_+_24069680 | 0.35 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr11_+_63022328 | 0.34 |
ENSMUST00000018361.10
|
Pmp22
|
peripheral myelin protein 22 |
| chr17_-_46343291 | 0.33 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr3_-_116762476 | 0.33 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr7_+_142030744 | 0.30 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr6_-_142647985 | 0.29 |
ENSMUST00000205202.3
ENSMUST00000073173.12 ENSMUST00000111771.8 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr7_-_44702269 | 0.27 |
ENSMUST00000057293.8
|
Prr12
|
proline rich 12 |
| chr3_-_75864195 | 0.27 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr4_-_126057263 | 0.27 |
ENSMUST00000097891.4
|
Sh3d21
|
SH3 domain containing 21 |
| chr4_-_126056412 | 0.26 |
ENSMUST00000094760.9
|
Sh3d21
|
SH3 domain containing 21 |
| chr10_-_37014859 | 0.24 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr9_-_65487322 | 0.23 |
ENSMUST00000068944.9
|
Plekho2
|
pleckstrin homology domain containing, family O member 2 |
| chr3_+_87878378 | 0.23 |
ENSMUST00000090973.12
|
Nes
|
nestin |
| chr6_-_124733441 | 0.22 |
ENSMUST00000088357.12
|
Atn1
|
atrophin 1 |
| chr3_+_87878436 | 0.22 |
ENSMUST00000160694.2
|
Nes
|
nestin |
| chr12_-_103304573 | 0.19 |
ENSMUST00000149431.2
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr16_+_18166045 | 0.18 |
ENSMUST00000239533.1
ENSMUST00000239534.1 |
ARVCF
|
armadillo repeat deleted in velocardiofacial syndrome |
| chr16_-_18165876 | 0.18 |
ENSMUST00000125287.9
|
Tango2
|
transport and golgi organization 2 |
| chr17_+_36176485 | 0.17 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_-_150868802 | 0.17 |
ENSMUST00000074783.12
ENSMUST00000137197.9 |
Hmcn1
|
hemicentin 1 |
| chr16_-_17540805 | 0.17 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr3_+_135144481 | 0.17 |
ENSMUST00000199582.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_142647944 | 0.16 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr11_+_69737437 | 0.16 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr7_-_99276310 | 0.15 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chrX_-_20797736 | 0.15 |
ENSMUST00000001156.8
|
Cfp
|
complement factor properdin |
| chr6_-_90758954 | 0.14 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr11_+_69737491 | 0.14 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr6_+_54016543 | 0.13 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr17_+_36176948 | 0.12 |
ENSMUST00000122899.8
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr17_-_36291087 | 0.12 |
ENSMUST00000055454.14
|
Prr3
|
proline-rich polypeptide 3 |
| chr12_+_80837284 | 0.12 |
ENSMUST00000220238.2
ENSMUST00000068519.7 |
Susd6
|
sushi domain containing 6 |
| chr7_+_28151370 | 0.12 |
ENSMUST00000190954.7
|
Lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr9_+_35332837 | 0.11 |
ENSMUST00000119129.10
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr10_-_5019044 | 0.11 |
ENSMUST00000095899.5
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr18_+_37847792 | 0.10 |
ENSMUST00000192511.2
ENSMUST00000193476.2 |
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr9_+_123307758 | 0.10 |
ENSMUST00000026269.4
|
Limd1
|
LIM domains containing 1 |
| chr1_-_173195236 | 0.08 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr11_-_96720738 | 0.07 |
ENSMUST00000107657.8
ENSMUST00000081775.12 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr3_+_135144287 | 0.07 |
ENSMUST00000196591.5
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_-_26080429 | 0.06 |
ENSMUST00000079461.15
ENSMUST00000176923.9 |
Wdr90
|
WD repeat domain 90 |
| chr4_-_94940425 | 0.06 |
ENSMUST00000107094.2
|
Jun
|
jun proto-oncogene |
| chr13_-_100240570 | 0.05 |
ENSMUST00000038104.12
|
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr5_-_146521629 | 0.04 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr8_-_69636825 | 0.04 |
ENSMUST00000185176.8
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr7_-_26928029 | 0.03 |
ENSMUST00000003850.8
|
Itpkc
|
inositol 1,4,5-trisphosphate 3-kinase C |
| chr13_-_104953370 | 0.02 |
ENSMUST00000022228.13
|
Cwc27
|
CWC27 spliceosome-associated protein |
| chr5_+_115769960 | 0.01 |
ENSMUST00000031492.15
|
Rab35
|
RAB35, member RAS oncogene family |
| chr13_-_104953631 | 0.01 |
ENSMUST00000154165.2
|
Cwc27
|
CWC27 spliceosome-associated protein |
| chr1_-_160079007 | 0.01 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr17_-_34834938 | 0.00 |
ENSMUST00000097345.10
ENSMUST00000015611.14 |
Egfl8
|
EGF-like domain 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0090076 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.7 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.3 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.4 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 2.6 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 4.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |