Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
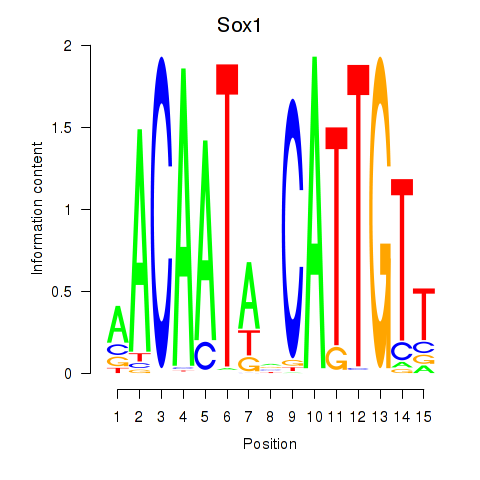
Results for Sox1
Z-value: 0.60
Transcription factors associated with Sox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox1
|
ENSMUSG00000096014.2 | SRY (sex determining region Y)-box 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox1 | mm39_v1_chr8_+_12445287_12445295 | 0.02 | 9.1e-01 | Click! |
Activity profile of Sox1 motif
Sorted Z-values of Sox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_13357892 | 7.48 |
ENSMUST00000108525.4
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chr19_-_8196196 | 2.89 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr17_-_34506744 | 2.60 |
ENSMUST00000174751.2
ENSMUST00000040655.14 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr17_+_34482183 | 2.57 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr6_+_37507108 | 1.94 |
ENSMUST00000040987.11
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr7_-_13856967 | 1.82 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr5_+_21391282 | 1.82 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr14_-_30645711 | 1.74 |
ENSMUST00000006697.17
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr7_+_67305162 | 1.74 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr5_+_21942139 | 1.51 |
ENSMUST00000030882.12
|
Pmpcb
|
peptidase (mitochondrial processing) beta |
| chr14_-_30645503 | 1.43 |
ENSMUST00000227995.2
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr10_-_125225298 | 1.30 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr15_-_74869684 | 0.97 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr13_+_42454922 | 0.89 |
ENSMUST00000021796.9
|
Edn1
|
endothelin 1 |
| chr15_-_74869483 | 0.86 |
ENSMUST00000023248.13
|
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr9_+_123921573 | 0.85 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr16_-_37474772 | 0.76 |
ENSMUST00000023514.4
|
Ndufb4
|
NADH:ubiquinone oxidoreductase subunit B4 |
| chr1_-_194813631 | 0.70 |
ENSMUST00000194111.6
ENSMUST00000193094.6 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr6_+_121160626 | 0.62 |
ENSMUST00000118234.8
ENSMUST00000088561.10 ENSMUST00000137432.8 ENSMUST00000120066.8 |
Pex26
|
peroxisomal biogenesis factor 26 |
| chr9_-_108183140 | 0.59 |
ENSMUST00000195615.2
|
Tcta
|
T cell leukemia translocation altered gene |
| chr12_+_52144511 | 0.53 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr8_+_13209141 | 0.53 |
ENSMUST00000033824.8
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr1_-_194813843 | 0.49 |
ENSMUST00000075451.12
ENSMUST00000191775.2 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr2_+_34296783 | 0.46 |
ENSMUST00000149383.8
ENSMUST00000124443.8 ENSMUST00000141253.2 ENSMUST00000113124.8 |
Mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr13_-_112788890 | 0.45 |
ENSMUST00000099166.10
|
Ddx4
|
DEAD box helicase 4 |
| chr2_-_84481058 | 0.39 |
ENSMUST00000111670.9
ENSMUST00000111697.9 ENSMUST00000111696.8 ENSMUST00000111678.8 ENSMUST00000111690.8 ENSMUST00000111695.8 ENSMUST00000111677.8 ENSMUST00000111698.8 ENSMUST00000099941.9 ENSMUST00000111676.8 ENSMUST00000111694.8 ENSMUST00000111675.8 ENSMUST00000111689.8 ENSMUST00000111687.8 ENSMUST00000111692.8 ENSMUST00000111685.8 ENSMUST00000111686.8 ENSMUST00000111688.8 ENSMUST00000111693.8 ENSMUST00000111684.8 |
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr13_-_112788829 | 0.39 |
ENSMUST00000075748.7
|
Ddx4
|
DEAD box helicase 4 |
| chr19_+_18818001 | 0.37 |
ENSMUST00000237020.2
|
Trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr7_+_79992839 | 0.36 |
ENSMUST00000032747.7
ENSMUST00000206480.2 ENSMUST00000206074.2 ENSMUST00000206122.2 |
Hddc3
|
HD domain containing 3 |
| chr4_-_120604445 | 0.35 |
ENSMUST00000030376.8
|
Kcnq4
|
potassium voltage-gated channel, subfamily Q, member 4 |
| chr2_-_84481101 | 0.34 |
ENSMUST00000111691.2
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr6_+_117818135 | 0.31 |
ENSMUST00000112859.8
ENSMUST00000137224.8 ENSMUST00000164472.8 ENSMUST00000112861.8 ENSMUST00000223041.2 |
Zfp637
|
zinc finger protein 637 |
| chr11_+_96024612 | 0.31 |
ENSMUST00000167258.8
|
Ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr6_+_114625771 | 0.26 |
ENSMUST00000182510.8
|
Atg7
|
autophagy related 7 |
| chr15_+_81511486 | 0.21 |
ENSMUST00000206833.2
|
Ep300
|
E1A binding protein p300 |
| chr11_+_96025045 | 0.20 |
ENSMUST00000107680.2
|
Ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr17_-_48003391 | 0.18 |
ENSMUST00000113300.8
|
Prickle4
|
prickle planar cell polarity protein 4 |
| chr10_+_84591919 | 0.16 |
ENSMUST00000060397.13
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr1_-_173703424 | 0.14 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr7_-_127490139 | 0.14 |
ENSMUST00000205300.2
ENSMUST00000121394.3 |
Prss53
|
protease, serine 53 |
| chr7_+_119289249 | 0.13 |
ENSMUST00000047045.10
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr7_-_46569617 | 0.13 |
ENSMUST00000210664.2
ENSMUST00000156335.9 |
Tsg101
|
tumor susceptibility gene 101 |
| chr7_-_103710652 | 0.12 |
ENSMUST00000074064.5
|
4930516K23Rik
|
RIKEN cDNA 4930516K23 gene |
| chr17_-_29162794 | 0.12 |
ENSMUST00000232977.2
|
Pxt1
|
peroxisomal, testis specific 1 |
| chr1_+_171723231 | 0.10 |
ENSMUST00000097466.3
|
Gm10521
|
predicted gene 10521 |
| chr18_+_37630044 | 0.10 |
ENSMUST00000059571.7
|
Pcdhb19
|
protocadherin beta 19 |
| chr16_+_8500694 | 0.09 |
ENSMUST00000202157.2
|
Gm5767
|
predicted gene 5767 |
| chr19_-_8382424 | 0.05 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr2_+_87185159 | 0.04 |
ENSMUST00000215163.3
|
Olfr1120
|
olfactory receptor 1120 |
| chr9_+_44238089 | 0.04 |
ENSMUST00000054708.5
|
Dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr5_-_129747129 | 0.02 |
ENSMUST00000049778.7
|
Zfp11
|
zinc finger protein 11 |
| chrX_+_111510223 | 0.01 |
ENSMUST00000113409.8
|
Zfp711
|
zinc finger protein 711 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.6 | 1.9 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.3 | 0.9 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 2.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 1.3 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.5 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 0.8 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 2.6 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.7 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 1.2 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 1.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.8 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.6 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:0014737 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) positive regulation of muscle atrophy(GO:0014737) |
| 0.1 | 3.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.3 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) amino acid homeostasis(GO:0080144) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 | 0.4 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.6 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.5 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.1 | 0.8 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 1.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.3 | GO:0050656 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.4 | 1.9 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.2 | 0.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 1.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 2.8 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 2.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.3 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 1.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |