Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
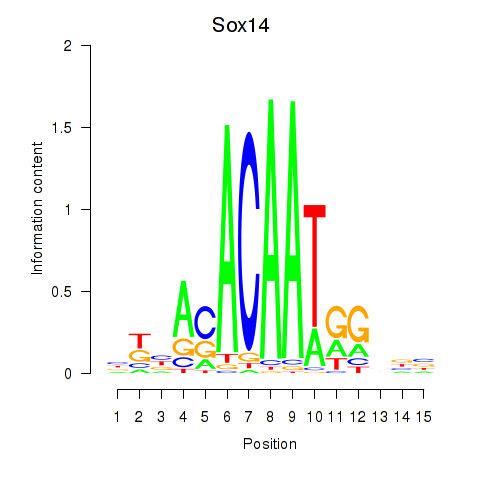
Results for Sox14
Z-value: 0.75
Transcription factors associated with Sox14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox14
|
ENSMUSG00000053747.10 | SRY (sex determining region Y)-box 14 |
Activity profile of Sox14 motif
Sorted Z-values of Sox14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142220553 | 5.69 |
ENSMUST00000105935.8
|
Igf2
|
insulin-like growth factor 2 |
| chr11_+_68936457 | 3.82 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr11_-_11987391 | 2.61 |
ENSMUST00000093321.12
|
Grb10
|
growth factor receptor bound protein 10 |
| chr1_+_134110142 | 2.26 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr15_+_73620213 | 2.18 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr17_-_36149100 | 2.06 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr19_+_44282113 | 1.98 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr2_+_120307390 | 1.96 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr17_-_36149142 | 1.89 |
ENSMUST00000001566.10
|
Tubb5
|
tubulin, beta 5 class I |
| chr7_+_78922947 | 1.81 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr1_-_128520002 | 1.80 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr7_+_24069680 | 1.78 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr2_+_119449192 | 1.75 |
ENSMUST00000028771.8
|
Nusap1
|
nucleolar and spindle associated protein 1 |
| chr14_+_30856687 | 1.74 |
ENSMUST00000090212.5
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr12_-_32111214 | 1.69 |
ENSMUST00000003079.12
ENSMUST00000036497.16 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chr11_+_58531220 | 1.65 |
ENSMUST00000075084.5
|
Trim58
|
tripartite motif-containing 58 |
| chr10_-_37014859 | 1.65 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chrX_-_165992145 | 1.58 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr13_-_55635851 | 1.54 |
ENSMUST00000109921.9
ENSMUST00000109923.9 ENSMUST00000021950.15 |
Dbn1
|
drebrin 1 |
| chr1_+_134109888 | 1.54 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr15_+_75027089 | 1.54 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr17_+_35278011 | 1.49 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr12_-_36092475 | 1.46 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr16_+_37597235 | 1.42 |
ENSMUST00000114763.3
|
Fstl1
|
follistatin-like 1 |
| chr7_+_121818692 | 1.41 |
ENSMUST00000033152.5
|
Chp2
|
calcineurin-like EF hand protein 2 |
| chr8_+_72889073 | 1.36 |
ENSMUST00000003575.11
|
Tpm4
|
tropomyosin 4 |
| chr2_-_151586063 | 1.32 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr8_+_72889607 | 1.31 |
ENSMUST00000238492.2
|
Tpm4
|
tropomyosin 4 |
| chr12_+_31315270 | 1.31 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chr9_-_103357564 | 1.30 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr7_+_99184858 | 1.29 |
ENSMUST00000032995.15
ENSMUST00000162404.8 |
Arrb1
|
arrestin, beta 1 |
| chr2_+_32766126 | 1.24 |
ENSMUST00000028135.15
|
Niban2
|
niban apoptosis regulator 2 |
| chr2_-_119448935 | 1.23 |
ENSMUST00000123818.2
|
Oip5
|
Opa interacting protein 5 |
| chr4_+_134195631 | 1.21 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr8_-_78451055 | 1.17 |
ENSMUST00000034029.8
|
Ednra
|
endothelin receptor type A |
| chrX_-_165992311 | 1.16 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chrX_+_161684563 | 1.05 |
ENSMUST00000112303.8
ENSMUST00000033727.14 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr17_-_18104182 | 1.05 |
ENSMUST00000061516.8
|
Fpr1
|
formyl peptide receptor 1 |
| chr13_+_112937385 | 1.03 |
ENSMUST00000070951.8
|
Plpp1
|
phospholipid phosphatase 1 |
| chr7_+_99184645 | 1.03 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr2_+_104420798 | 1.03 |
ENSMUST00000028599.8
|
Cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr4_-_6990774 | 1.03 |
ENSMUST00000039987.4
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr10_-_117212860 | 1.02 |
ENSMUST00000069168.13
ENSMUST00000176686.8 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr13_+_112937320 | 1.01 |
ENSMUST00000016144.12
|
Plpp1
|
phospholipid phosphatase 1 |
| chr10_-_117212826 | 1.00 |
ENSMUST00000177145.8
ENSMUST00000176670.8 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr1_-_43203051 | 0.98 |
ENSMUST00000008280.14
|
Fhl2
|
four and a half LIM domains 2 |
| chr1_-_72914036 | 0.97 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr1_-_79838897 | 0.95 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr12_+_31315227 | 0.94 |
ENSMUST00000169088.8
|
Lamb1
|
laminin B1 |
| chr14_-_60434622 | 0.94 |
ENSMUST00000131670.3
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr12_+_108572015 | 0.93 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chrX_+_92698469 | 0.92 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr10_+_96453408 | 0.92 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr9_+_98305014 | 0.91 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chr11_-_79145489 | 0.91 |
ENSMUST00000017821.12
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr4_+_126915104 | 0.90 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr10_+_39245746 | 0.90 |
ENSMUST00000063091.13
ENSMUST00000099967.10 ENSMUST00000126486.8 |
Fyn
|
Fyn proto-oncogene |
| chr5_+_142946098 | 0.88 |
ENSMUST00000031565.15
ENSMUST00000198017.5 |
Fscn1
|
fascin actin-bundling protein 1 |
| chr19_-_10847121 | 0.85 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr1_-_180641159 | 0.85 |
ENSMUST00000162118.8
ENSMUST00000159685.2 ENSMUST00000161308.8 |
H3f3a
|
H3.3 histone A |
| chr17_-_79662514 | 0.84 |
ENSMUST00000068958.9
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr9_+_95519654 | 0.83 |
ENSMUST00000015498.9
|
Pcolce2
|
procollagen C-endopeptidase enhancer 2 |
| chr7_-_100164007 | 0.83 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chrX_+_161684735 | 0.83 |
ENSMUST00000112302.8
ENSMUST00000112301.8 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr1_+_39940189 | 0.81 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr15_-_79430742 | 0.81 |
ENSMUST00000231053.2
ENSMUST00000229431.2 |
Ddx17
|
DEAD box helicase 17 |
| chr16_+_90182895 | 0.79 |
ENSMUST00000065856.8
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr1_-_180641430 | 0.79 |
ENSMUST00000162814.8
|
H3f3a
|
H3.3 histone A |
| chr15_+_88635852 | 0.77 |
ENSMUST00000041297.15
|
Zbed4
|
zinc finger, BED type containing 4 |
| chr1_-_180641099 | 0.76 |
ENSMUST00000159789.2
ENSMUST00000081026.11 |
H3f3a
|
H3.3 histone A |
| chr5_+_142946598 | 0.76 |
ENSMUST00000129306.4
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr17_+_48623157 | 0.75 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chrX_+_133486391 | 0.74 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr1_+_90131702 | 0.72 |
ENSMUST00000065587.5
ENSMUST00000159654.2 |
Ackr3
|
atypical chemokine receptor 3 |
| chr10_-_91007387 | 0.72 |
ENSMUST00000099355.12
ENSMUST00000105293.11 ENSMUST00000092219.14 ENSMUST00000020123.7 ENSMUST00000072239.14 |
Tmpo
|
thymopoietin |
| chr1_+_135945705 | 0.72 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr3_+_7568481 | 0.72 |
ENSMUST00000051064.9
ENSMUST00000193010.2 |
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr3_-_37778470 | 0.71 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr1_+_82817388 | 0.70 |
ENSMUST00000190052.7
ENSMUST00000063380.11 ENSMUST00000187899.7 ENSMUST00000186302.7 ENSMUST00000190046.7 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr5_+_90708962 | 0.70 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr1_+_135768595 | 0.70 |
ENSMUST00000112087.9
ENSMUST00000178854.8 ENSMUST00000027671.12 ENSMUST00000179863.8 ENSMUST00000112085.9 ENSMUST00000112086.3 |
Tnnt2
|
troponin T2, cardiac |
| chr12_+_116369017 | 0.70 |
ENSMUST00000084828.5
ENSMUST00000222469.2 ENSMUST00000221114.2 ENSMUST00000221970.2 |
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr4_+_109272828 | 0.69 |
ENSMUST00000106618.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr10_+_126899468 | 0.69 |
ENSMUST00000120226.8
ENSMUST00000133115.8 |
Cdk4
|
cyclin-dependent kinase 4 |
| chr11_-_102338473 | 0.68 |
ENSMUST00000049057.5
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr11_+_23206001 | 0.68 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr3_+_89986831 | 0.64 |
ENSMUST00000029549.16
|
Tpm3
|
tropomyosin 3, gamma |
| chr6_-_97594498 | 0.64 |
ENSMUST00000113355.9
|
Frmd4b
|
FERM domain containing 4B |
| chr1_+_118317153 | 0.63 |
ENSMUST00000189738.7
ENSMUST00000187713.7 ENSMUST00000165223.8 ENSMUST00000189570.7 ENSMUST00000191445.7 ENSMUST00000189262.7 |
Clasp1
|
CLIP associating protein 1 |
| chr4_-_136329953 | 0.63 |
ENSMUST00000105847.8
ENSMUST00000116273.9 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr16_+_23044763 | 0.61 |
ENSMUST00000178797.8
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr9_-_66032134 | 0.60 |
ENSMUST00000034946.15
|
Snx1
|
sorting nexin 1 |
| chr11_+_60590498 | 0.60 |
ENSMUST00000052346.10
|
Llgl1
|
LLGL1 scribble cell polarity complex component |
| chr11_-_106606076 | 0.60 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr10_-_6930376 | 0.59 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_+_89986925 | 0.59 |
ENSMUST00000118566.8
ENSMUST00000119158.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr19_-_47907705 | 0.59 |
ENSMUST00000095998.7
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr3_+_60503051 | 0.58 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr9_-_88364593 | 0.58 |
ENSMUST00000173801.8
ENSMUST00000069221.12 ENSMUST00000172508.2 |
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr17_-_48474356 | 0.57 |
ENSMUST00000027764.10
ENSMUST00000053612.14 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr4_-_129334593 | 0.57 |
ENSMUST00000053042.6
ENSMUST00000106046.8 |
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr15_-_103242697 | 0.56 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr14_-_51295099 | 0.55 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr18_+_36661198 | 0.54 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr14_-_52553764 | 0.54 |
ENSMUST00000135523.5
|
Sall2
|
spalt like transcription factor 2 |
| chr3_+_151143557 | 0.53 |
ENSMUST00000196970.3
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr19_-_46321218 | 0.53 |
ENSMUST00000238062.2
|
Cuedc2
|
CUE domain containing 2 |
| chr6_-_128765529 | 0.52 |
ENSMUST00000167691.9
ENSMUST00000174404.8 |
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr9_+_22010512 | 0.52 |
ENSMUST00000214601.2
ENSMUST00000001384.6 |
Cnn1
|
calponin 1 |
| chr2_-_60793536 | 0.52 |
ENSMUST00000028347.13
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr1_+_135945798 | 0.52 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr5_-_137869969 | 0.51 |
ENSMUST00000196162.5
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chrX_-_9983836 | 0.50 |
ENSMUST00000115543.3
ENSMUST00000044789.10 ENSMUST00000115544.9 |
Srpx
|
sushi-repeat-containing protein |
| chr11_+_109376432 | 0.50 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr5_+_145217272 | 0.50 |
ENSMUST00000200246.2
|
Zscan25
|
zinc finger and SCAN domain containing 25 |
| chr14_-_72840373 | 0.49 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr3_+_40978804 | 0.49 |
ENSMUST00000099121.10
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr10_-_79710468 | 0.48 |
ENSMUST00000092325.11
|
Plppr3
|
phospholipid phosphatase related 3 |
| chr17_-_31363245 | 0.46 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr11_-_68848271 | 0.46 |
ENSMUST00000108671.2
|
Arhgef15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr1_-_173703424 | 0.45 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr15_+_81469538 | 0.45 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr2_-_60711706 | 0.45 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr12_-_55033130 | 0.44 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr5_+_115039359 | 0.44 |
ENSMUST00000124716.3
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr9_+_13677266 | 0.43 |
ENSMUST00000152532.8
|
Mtmr2
|
myotubularin related protein 2 |
| chr5_+_108213608 | 0.43 |
ENSMUST00000081567.11
ENSMUST00000170319.8 ENSMUST00000112626.8 |
Mtf2
|
metal response element binding transcription factor 2 |
| chr3_-_104960264 | 0.43 |
ENSMUST00000098763.7
ENSMUST00000197437.5 |
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr2_-_102016717 | 0.43 |
ENSMUST00000058790.12
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr11_-_106605772 | 0.41 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr1_-_16174387 | 0.41 |
ENSMUST00000149566.2
|
Rpl7
|
ribosomal protein L7 |
| chrX_-_100777806 | 0.41 |
ENSMUST00000056614.7
|
Cxcr3
|
chemokine (C-X-C motif) receptor 3 |
| chr16_+_4867876 | 0.40 |
ENSMUST00000230703.2
ENSMUST00000052449.6 |
Ubn1
|
ubinuclein 1 |
| chr13_-_22016364 | 0.40 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr2_-_102016665 | 0.40 |
ENSMUST00000111222.2
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr12_-_99849660 | 0.40 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr15_-_79430942 | 0.40 |
ENSMUST00000054014.9
ENSMUST00000229877.2 |
Ddx17
|
DEAD box helicase 17 |
| chr3_-_103553347 | 0.40 |
ENSMUST00000117271.3
|
Atg4a-ps
|
autophagy related 4A, pseudogene |
| chr8_+_106785434 | 0.39 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr11_-_5049223 | 0.39 |
ENSMUST00000079949.13
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr3_+_129695068 | 0.38 |
ENSMUST00000029626.9
|
Casp6
|
caspase 6 |
| chr3_-_104960437 | 0.38 |
ENSMUST00000077548.12
|
Cttnbp2nl
|
CTTNBP2 N-terminal like |
| chr10_-_125164826 | 0.37 |
ENSMUST00000211781.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr1_-_135302971 | 0.37 |
ENSMUST00000041240.4
|
Shisa4
|
shisa family member 4 |
| chr6_+_120341055 | 0.36 |
ENSMUST00000005108.10
|
Kdm5a
|
lysine (K)-specific demethylase 5A |
| chr7_-_44635813 | 0.36 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr2_-_154411765 | 0.36 |
ENSMUST00000103145.11
|
E2f1
|
E2F transcription factor 1 |
| chrX_+_137986975 | 0.36 |
ENSMUST00000033625.2
|
4930513O06Rik
|
RIKEN cDNA 4930513O06 gene |
| chr4_+_101504938 | 0.35 |
ENSMUST00000106927.2
|
Leprot
|
leptin receptor overlapping transcript |
| chr2_+_3115250 | 0.35 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr4_+_101504909 | 0.35 |
ENSMUST00000030254.15
|
Leprot
|
leptin receptor overlapping transcript |
| chr1_+_192855776 | 0.35 |
ENSMUST00000161235.3
ENSMUST00000160077.2 ENSMUST00000178744.2 ENSMUST00000192189.2 ENSMUST00000110831.4 ENSMUST00000191613.2 |
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
| chr9_+_65102635 | 0.34 |
ENSMUST00000216702.2
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr15_+_99600149 | 0.34 |
ENSMUST00000229236.2
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr3_+_151143524 | 0.33 |
ENSMUST00000046977.12
|
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr12_-_115122455 | 0.33 |
ENSMUST00000103523.2
|
Ighv1-53
|
immunoglobulin heavy variable 1-53 |
| chr1_+_9868332 | 0.32 |
ENSMUST00000166384.8
ENSMUST00000168907.8 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr6_+_37847721 | 0.32 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr12_+_80837284 | 0.32 |
ENSMUST00000220238.2
ENSMUST00000068519.7 |
Susd6
|
sushi domain containing 6 |
| chr13_+_44994167 | 0.32 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr1_+_82817794 | 0.32 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr15_-_96540760 | 0.31 |
ENSMUST00000088452.11
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr7_+_24230063 | 0.31 |
ENSMUST00000049020.9
|
Irgq
|
immunity-related GTPase family, Q |
| chr17_+_34457868 | 0.31 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr12_-_55033113 | 0.30 |
ENSMUST00000038926.13
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr8_+_11778039 | 0.30 |
ENSMUST00000110909.9
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr3_+_61269059 | 0.30 |
ENSMUST00000049064.4
|
Rap2b
|
RAP2B, member of RAS oncogene family |
| chr19_-_33567708 | 0.30 |
ENSMUST00000112508.9
|
Lipo3
|
lipase, member O3 |
| chr5_-_33432310 | 0.30 |
ENSMUST00000201372.3
ENSMUST00000202962.4 ENSMUST00000201575.4 ENSMUST00000202868.4 ENSMUST00000079746.10 |
Ctbp1
|
C-terminal binding protein 1 |
| chrX_+_100473161 | 0.29 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr1_-_77491683 | 0.29 |
ENSMUST00000186930.2
ENSMUST00000027451.13 ENSMUST00000188797.7 |
Epha4
|
Eph receptor A4 |
| chr6_+_122490534 | 0.29 |
ENSMUST00000032210.14
ENSMUST00000148517.8 |
Mfap5
|
microfibrillar associated protein 5 |
| chr5_+_139197689 | 0.29 |
ENSMUST00000148772.8
ENSMUST00000110882.8 |
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr2_-_163486998 | 0.29 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr13_+_49495025 | 0.28 |
ENSMUST00000048544.14
ENSMUST00000110084.4 ENSMUST00000110085.11 |
Bicd2
|
BICD cargo adaptor 2 |
| chr3_+_7431717 | 0.27 |
ENSMUST00000192468.2
ENSMUST00000028999.12 |
Pkia
|
protein kinase inhibitor, alpha |
| chr10_-_18890281 | 0.27 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr13_+_21995906 | 0.26 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17 |
| chr2_-_131953359 | 0.26 |
ENSMUST00000128899.2
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr19_-_57107413 | 0.26 |
ENSMUST00000111528.8
ENSMUST00000111529.8 ENSMUST00000104902.9 |
Ablim1
|
actin-binding LIM protein 1 |
| chr13_-_76091931 | 0.25 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr12_-_108801763 | 0.25 |
ENSMUST00000021693.4
|
Slc25a29
|
solute carrier family 25 (mitochondrial carrier, palmitoylcarnitine transporter), member 29 |
| chr14_+_75373766 | 0.25 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr1_+_88154727 | 0.25 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr6_-_42670021 | 0.25 |
ENSMUST00000121083.8
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr6_-_82751429 | 0.24 |
ENSMUST00000000642.11
|
Hk2
|
hexokinase 2 |
| chr7_+_89053562 | 0.24 |
ENSMUST00000058755.5
|
Fzd4
|
frizzled class receptor 4 |
| chr4_-_132149704 | 0.23 |
ENSMUST00000152271.8
ENSMUST00000084170.12 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr17_-_36014892 | 0.23 |
ENSMUST00000097333.10
ENSMUST00000003628.13 |
Ddr1
|
discoidin domain receptor family, member 1 |
| chr17_+_24633614 | 0.23 |
ENSMUST00000115371.9
ENSMUST00000088512.13 ENSMUST00000163717.2 |
Rnps1
|
RNA binding protein with serine rich domain 1 |
| chr2_-_154411640 | 0.23 |
ENSMUST00000000894.6
|
E2f1
|
E2F transcription factor 1 |
| chr7_+_79675727 | 0.23 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr11_+_96209093 | 0.22 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr6_+_51447613 | 0.22 |
ENSMUST00000114445.8
ENSMUST00000114446.8 ENSMUST00000141711.3 |
Cbx3
|
chromobox 3 |
| chrX_+_97979920 | 0.22 |
ENSMUST00000113811.8
|
Yipf6
|
Yip1 domain family, member 6 |
| chr12_-_21467437 | 0.22 |
ENSMUST00000103002.8
ENSMUST00000155480.9 ENSMUST00000135088.9 |
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr14_+_69585036 | 0.22 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr13_-_58261406 | 0.21 |
ENSMUST00000160860.9
|
Klhl3
|
kelch-like 3 |
| chr10_+_23770586 | 0.21 |
ENSMUST00000041416.8
|
Vnn1
|
vanin 1 |
| chr5_-_115410941 | 0.21 |
ENSMUST00000040555.15
ENSMUST00000112096.9 ENSMUST00000112097.8 |
Rnf10
|
ring finger protein 10 |
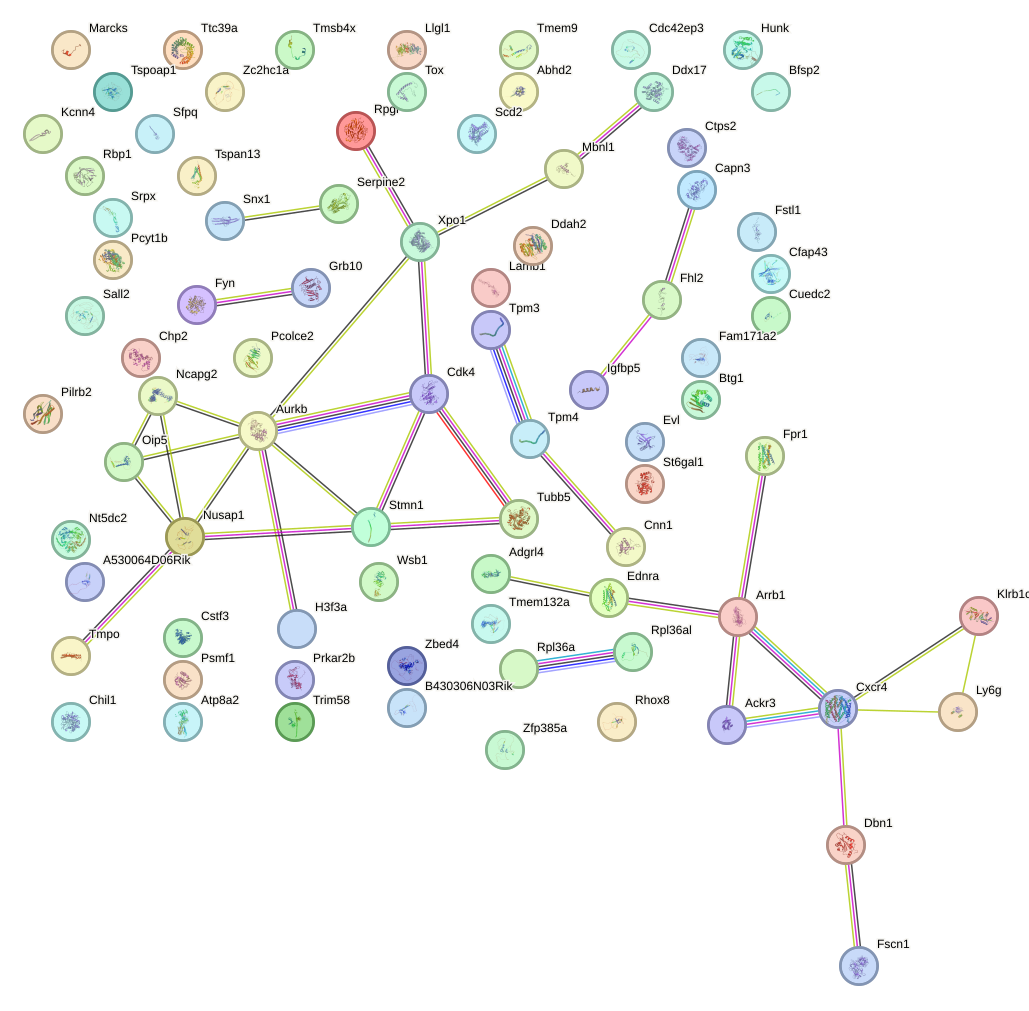
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.6 | 2.2 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.5 | 1.5 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.5 | 2.0 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.5 | 2.4 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.5 | 5.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.4 | 1.8 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.3 | 2.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.0 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.3 | 2.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 1.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.3 | 0.9 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.3 | 2.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 2.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 1.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.2 | 0.8 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 1.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 0.6 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 1.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.4 | GO:0014737 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) positive regulation of muscle atrophy(GO:0014737) |
| 0.1 | 0.6 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 3.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.7 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 2.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.5 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.5 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.9 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.6 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.1 | 2.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.9 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 1.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.9 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 1.2 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.7 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.1 | 0.3 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 2.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.5 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 1.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 1.8 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.3 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 1.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.2 | GO:1902022 | mitochondrial ornithine transport(GO:0000066) L-lysine transport(GO:1902022) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) L-lysine transmembrane transport(GO:1903401) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.4 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.9 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.5 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 2.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 1.0 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.6 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.3 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.2 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 | 0.4 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.2 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 1.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 1.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.3 | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 4.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.4 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.0 | 1.7 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.6 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.0 | 0.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0060369 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.3 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.6 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.6 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.1 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.6 | 3.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.6 | 2.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.5 | 4.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 1.5 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.3 | 1.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.3 | 1.6 | GO:0044393 | microspike(GO:0044393) |
| 0.3 | 3.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 3.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 0.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 2.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) GAIT complex(GO:0097452) |
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.2 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 3.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.5 | 1.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.4 | 1.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 1.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.3 | 2.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.3 | 3.8 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 1.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 2.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 0.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 4.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 1.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 1.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 3.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 2.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 1.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 0.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 2.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 5.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 2.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.3 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.2 | GO:0005289 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.1 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 1.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 3.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.2 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 2.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 2.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.5 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.9 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 7.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.3 | 6.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 3.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 2.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |