Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
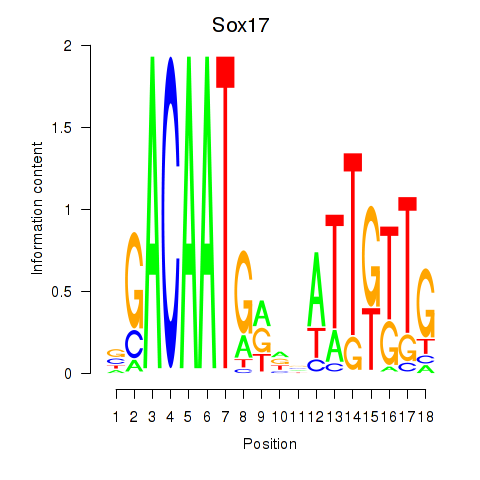
Results for Sox17
Z-value: 1.06
Transcription factors associated with Sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox17
|
ENSMUSG00000025902.14 | SRY (sex determining region Y)-box 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox17 | mm39_v1_chr1_-_4566619_4566643 | -0.61 | 8.0e-05 | Click! |
Activity profile of Sox17 motif
Sorted Z-values of Sox17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_48516447 | 19.86 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr8_-_93956143 | 9.08 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr2_+_102489558 | 9.05 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr16_+_22769822 | 8.66 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr16_-_18232202 | 8.60 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr16_+_22769844 | 8.10 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr8_-_94063823 | 8.05 |
ENSMUST00000044602.8
|
Ces1g
|
carboxylesterase 1G |
| chr8_-_93924426 | 8.01 |
ENSMUST00000034172.8
|
Ces1d
|
carboxylesterase 1D |
| chr6_-_128503666 | 6.84 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chrX_+_59044796 | 6.39 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr3_-_98670369 | 5.90 |
ENSMUST00000107019.8
ENSMUST00000107018.8 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr8_+_107877252 | 5.65 |
ENSMUST00000034400.5
|
Cyb5b
|
cytochrome b5 type B |
| chr5_-_89605622 | 5.56 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr7_-_100306160 | 5.24 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr12_-_84447702 | 4.67 |
ENSMUST00000122194.8
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr5_-_104169696 | 4.46 |
ENSMUST00000119025.2
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr12_-_84447625 | 4.16 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr1_-_162694076 | 4.12 |
ENSMUST00000046049.14
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr5_+_90708962 | 3.99 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr4_-_42773987 | 3.97 |
ENSMUST00000095114.5
|
Ccl21a
|
chemokine (C-C motif) ligand 21A (serine) |
| chr9_+_57428490 | 3.72 |
ENSMUST00000000090.8
|
Cox5a
|
cytochrome c oxidase subunit 5A |
| chr5_-_53370761 | 3.72 |
ENSMUST00000031090.8
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr6_+_121709891 | 3.05 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr11_-_59340739 | 2.64 |
ENSMUST00000136436.2
ENSMUST00000150297.2 ENSMUST00000010038.10 ENSMUST00000156146.8 ENSMUST00000132969.8 ENSMUST00000120940.8 |
Snap47
|
synaptosomal-associated protein, 47 |
| chr8_-_41507808 | 2.59 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr17_-_24428351 | 2.55 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr7_+_28937859 | 2.44 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr13_-_35211060 | 2.33 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr16_-_22847808 | 2.21 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr5_-_38659449 | 2.20 |
ENSMUST00000005238.13
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr16_-_22847760 | 2.08 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr11_-_109986763 | 2.05 |
ENSMUST00000046223.14
ENSMUST00000106664.10 ENSMUST00000106662.2 |
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr11_-_77784922 | 2.04 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr16_-_22847829 | 2.03 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr11_-_109986804 | 1.97 |
ENSMUST00000100287.9
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a |
| chr19_+_8897732 | 1.95 |
ENSMUST00000096243.7
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr19_+_34560922 | 1.92 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr7_-_127490139 | 1.89 |
ENSMUST00000205300.2
ENSMUST00000121394.3 |
Prss53
|
protease, serine 53 |
| chr1_-_182169150 | 1.84 |
ENSMUST00000051431.10
|
Fbxo28
|
F-box protein 28 |
| chr10_+_80084955 | 1.81 |
ENSMUST00000105364.8
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr4_+_41903610 | 1.79 |
ENSMUST00000098128.4
|
Ccl21d
|
chemokine (C-C motif) ligand 21D |
| chr9_+_50405817 | 1.78 |
ENSMUST00000114474.8
ENSMUST00000188047.2 |
Plet1
|
placenta expressed transcript 1 |
| chr4_+_42612121 | 1.75 |
ENSMUST00000178168.3
|
Gm10591
|
predicted gene 10591 |
| chr4_+_42255693 | 1.68 |
ENSMUST00000178864.3
|
Ccl21b
|
chemokine (C-C motif) ligand 21B (leucine) |
| chr7_+_28937898 | 1.66 |
ENSMUST00000138128.3
ENSMUST00000142519.3 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr2_+_80447389 | 1.65 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr8_+_118428643 | 1.64 |
ENSMUST00000034304.9
|
Hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr16_+_8288627 | 1.55 |
ENSMUST00000046470.16
ENSMUST00000150790.2 ENSMUST00000142899.2 |
Mettl22
|
methyltransferase like 22 |
| chr16_+_22877000 | 1.55 |
ENSMUST00000039492.14
ENSMUST00000023589.15 ENSMUST00000089902.8 |
Kng1
|
kininogen 1 |
| chr17_+_47747540 | 1.52 |
ENSMUST00000037701.13
|
AI661453
|
expressed sequence AI661453 |
| chr17_+_47747657 | 1.52 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr7_+_28937746 | 1.50 |
ENSMUST00000108238.8
ENSMUST00000032809.10 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr1_+_78488499 | 1.45 |
ENSMUST00000012331.7
|
Mogat1
|
monoacylglycerol O-acyltransferase 1 |
| chr19_-_32717166 | 1.39 |
ENSMUST00000235142.2
ENSMUST00000070210.6 ENSMUST00000236011.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chrX_-_47966588 | 1.38 |
ENSMUST00000114928.2
|
Fsip2l
|
fibrous sheath-interacting protein 2-like |
| chr13_+_19374502 | 1.35 |
ENSMUST00000198330.2
ENSMUST00000103555.3 |
Trgv6
|
T cell receptor gamma, variable 6 |
| chr19_-_32717138 | 1.34 |
ENSMUST00000236985.2
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr9_+_108269992 | 1.34 |
ENSMUST00000192995.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chr17_+_8529932 | 1.32 |
ENSMUST00000154553.2
ENSMUST00000140890.3 |
Sft2d1
Gm49987
|
SFT2 domain containing 1 predicted gene, 49987 |
| chr11_-_68291638 | 1.29 |
ENSMUST00000108674.9
|
Ntn1
|
netrin 1 |
| chr5_-_38659422 | 1.28 |
ENSMUST00000147664.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr16_-_22848153 | 1.26 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr5_-_125466240 | 1.22 |
ENSMUST00000108707.3
|
Ubc
|
ubiquitin C |
| chr9_+_66853343 | 1.22 |
ENSMUST00000040917.14
ENSMUST00000127896.8 |
Rps27l
|
ribosomal protein S27-like |
| chr11_-_86698484 | 1.20 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr2_+_52747855 | 1.18 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr13_-_74642055 | 1.15 |
ENSMUST00000202645.4
ENSMUST00000221173.2 |
Zfp825
|
zinc finger protein 825 |
| chr1_-_60605867 | 1.07 |
ENSMUST00000027168.12
ENSMUST00000090293.11 ENSMUST00000140485.8 |
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr9_+_3335507 | 1.06 |
ENSMUST00000212154.2
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr2_-_57003064 | 1.04 |
ENSMUST00000112627.2
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr7_+_97049210 | 0.96 |
ENSMUST00000032882.9
ENSMUST00000149122.2 |
Ndufc2
|
NADH:ubiquinone oxidoreductase subunit C2 |
| chr17_-_40553176 | 0.94 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr7_-_28329924 | 0.94 |
ENSMUST00000159095.2
ENSMUST00000159418.8 ENSMUST00000159560.3 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr9_+_108270020 | 0.91 |
ENSMUST00000035234.6
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene |
| chrY_-_26732144 | 0.90 |
ENSMUST00000189518.2
|
Gm20890
|
predicted gene, 20890 |
| chrY_-_27477265 | 0.90 |
ENSMUST00000188744.2
|
Gm21488
|
predicted gene, 21488 |
| chr19_+_6356486 | 0.89 |
ENSMUST00000025681.8
|
Cdc42bpg
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chrY_-_28226857 | 0.89 |
ENSMUST00000188864.2
|
Gm20894
|
predicted gene, 20894 |
| chr18_+_80298441 | 0.86 |
ENSMUST00000123750.8
|
Slc66a2
|
solute carrier family 66 member 2 |
| chr9_-_49338321 | 0.85 |
ENSMUST00000034792.7
|
Ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr13_-_99584091 | 0.79 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
| chr1_+_171330978 | 0.78 |
ENSMUST00000081527.2
|
Alyref2
|
Aly/REF export factor 2 |
| chr19_+_6234392 | 0.78 |
ENSMUST00000025699.9
ENSMUST00000113528.2 |
Majin
|
membrane anchored junction protein |
| chr12_-_40249314 | 0.72 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr7_-_28330322 | 0.67 |
ENSMUST00000040112.5
ENSMUST00000239470.2 |
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr5_-_74863514 | 0.66 |
ENSMUST00000117388.8
|
Lnx1
|
ligand of numb-protein X 1 |
| chr18_+_37440497 | 0.66 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr6_-_112364974 | 0.62 |
ENSMUST00000238755.2
ENSMUST00000060847.6 |
Ssu2
|
ssu-2 homolog (C. elegans) |
| chr8_+_4298189 | 0.60 |
ENSMUST00000110993.2
|
Tgfbr3l
|
transforming growth factor, beta receptor III-like |
| chr6_-_144155197 | 0.59 |
ENSMUST00000038815.14
ENSMUST00000111749.8 ENSMUST00000170367.9 |
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr8_-_25215856 | 0.57 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr13_+_54225828 | 0.56 |
ENSMUST00000021930.10
|
Sfxn1
|
sideroflexin 1 |
| chrX_+_73171069 | 0.54 |
ENSMUST00000033771.11
ENSMUST00000101457.10 |
Opn1mw
|
opsin 1 (cone pigments), medium-wave-sensitive (color blindness, deutan) |
| chr14_+_54664359 | 0.53 |
ENSMUST00000010550.12
ENSMUST00000199195.3 ENSMUST00000196273.2 |
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr9_+_110663656 | 0.53 |
ENSMUST00000011391.12
ENSMUST00000146794.4 |
Prss45
|
protease, serine 45 |
| chr18_-_33346885 | 0.52 |
ENSMUST00000025236.9
|
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr10_+_77891161 | 0.50 |
ENSMUST00000131825.8
ENSMUST00000139539.8 ENSMUST00000123940.2 |
Dnmt3l
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr10_-_62198043 | 0.50 |
ENSMUST00000149534.2
|
Hk1
|
hexokinase 1 |
| chr1_-_38875757 | 0.50 |
ENSMUST00000147695.9
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr2_-_76698725 | 0.48 |
ENSMUST00000149616.8
ENSMUST00000152185.8 ENSMUST00000130915.8 ENSMUST00000155365.8 ENSMUST00000128071.8 |
Ttn
|
titin |
| chr3_-_107240989 | 0.47 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr10_-_128012336 | 0.45 |
ENSMUST00000092033.4
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr7_-_127897253 | 0.45 |
ENSMUST00000033044.16
|
Rusf1
|
RUS family member 1 |
| chr6_-_144155167 | 0.45 |
ENSMUST00000077160.12
|
Sox5
|
SRY (sex determining region Y)-box 5 |
| chr13_-_27852017 | 0.44 |
ENSMUST00000006660.7
|
Prl7a2
|
prolactin family 7, subfamily a, member 2 |
| chr7_+_16693604 | 0.43 |
ENSMUST00000038163.8
|
Pnmal1
|
PNMA-like 1 |
| chr5_-_107745404 | 0.40 |
ENSMUST00000124140.2
|
Glmn
|
glomulin, FKBP associated protein |
| chr10_-_23226684 | 0.39 |
ENSMUST00000220299.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr14_-_121935829 | 0.38 |
ENSMUST00000040700.9
ENSMUST00000212181.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr5_+_27022355 | 0.38 |
ENSMUST00000071500.13
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr7_+_119927885 | 0.31 |
ENSMUST00000207220.2
ENSMUST00000121265.7 ENSMUST00000076272.5 |
Abca15
|
ATP-binding cassette, sub-family A (ABC1), member 15 |
| chr17_+_48070100 | 0.30 |
ENSMUST00000125177.2
|
Tfeb
|
transcription factor EB |
| chr6_+_116466052 | 0.29 |
ENSMUST00000203700.3
|
Olfr211
|
olfactory receptor 211 |
| chr2_-_121287174 | 0.27 |
ENSMUST00000110613.9
ENSMUST00000056312.10 |
Serinc4
|
serine incorporator 4 |
| chr2_+_89804937 | 0.26 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr19_+_12394797 | 0.26 |
ENSMUST00000216506.2
ENSMUST00000214153.2 |
Olfr1441
|
olfactory receptor 1441 |
| chr10_-_128904846 | 0.26 |
ENSMUST00000204515.3
|
Olfr766-ps1
|
olfactory receptor 766, pseudogene 1 |
| chr11_-_97666701 | 0.25 |
ENSMUST00000107576.2
|
1700001P01Rik
|
RIKEN cDNA 1700001P01 gene |
| chr7_-_139785877 | 0.25 |
ENSMUST00000215815.2
|
Olfr524
|
olfactory receptor 524 |
| chr7_+_103583554 | 0.23 |
ENSMUST00000214711.2
|
Olfr632
|
olfactory receptor 632 |
| chr17_+_46739852 | 0.23 |
ENSMUST00000113465.10
ENSMUST00000024764.12 ENSMUST00000165993.2 |
Crip3
|
cysteine-rich protein 3 |
| chr2_-_151528259 | 0.22 |
ENSMUST00000149319.2
|
Tmem74bos
|
transmembrane 74B, opposite strand |
| chr7_+_99384352 | 0.21 |
ENSMUST00000098264.2
|
Olfr520
|
olfactory receptor 520 |
| chr10_-_102864921 | 0.21 |
ENSMUST00000217946.2
ENSMUST00000219194.2 |
Alx1
|
ALX homeobox 1 |
| chr18_-_33346819 | 0.20 |
ENSMUST00000119991.8
ENSMUST00000118990.2 |
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr11_+_98537582 | 0.18 |
ENSMUST00000093938.10
ENSMUST00000017355.12 |
Gsdma2
|
gasdermin A2 |
| chr8_-_25215778 | 0.18 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr6_+_121222865 | 0.14 |
ENSMUST00000032198.11
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr8_+_106154266 | 0.13 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr6_-_129761233 | 0.13 |
ENSMUST00000118532.8
|
Klrh1
|
killer cell lectin-like receptor subfamily H, member 1 |
| chr4_+_118818775 | 0.13 |
ENSMUST00000058651.5
|
Lao1
|
L-amino acid oxidase 1 |
| chr3_+_92259448 | 0.12 |
ENSMUST00000068399.2
|
Sprr2e
|
small proline-rich protein 2E |
| chr11_-_49005701 | 0.11 |
ENSMUST00000060398.3
ENSMUST00000215553.2 ENSMUST00000109201.2 |
Olfr1396
|
olfactory receptor 1396 |
| chr9_-_35701371 | 0.11 |
ENSMUST00000034619.2
|
Pate6
|
prostate and testis expressed 6 |
| chr13_+_33668761 | 0.10 |
ENSMUST00000081927.4
|
Serpinb9g
|
serine (or cysteine) peptidase inhibitor, clade B, member 9g |
| chr17_-_23783063 | 0.06 |
ENSMUST00000095606.5
|
Zfp213
|
zinc finger protein 213 |
| chr5_+_138185747 | 0.06 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr6_+_24857967 | 0.06 |
ENSMUST00000200968.4
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr6_+_68414401 | 0.05 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr3_-_87934772 | 0.04 |
ENSMUST00000005014.9
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr9_-_75504926 | 0.04 |
ENSMUST00000164100.2
|
Tmod2
|
tropomodulin 2 |
| chr14_+_68321302 | 0.04 |
ENSMUST00000022639.8
|
Nefl
|
neurofilament, light polypeptide |
| chr7_+_44984723 | 0.03 |
ENSMUST00000211327.2
|
Hrc
|
histidine rich calcium binding protein |
| chr3_+_132335575 | 0.03 |
ENSMUST00000212804.2
ENSMUST00000212852.2 |
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr5_-_5713264 | 0.03 |
ENSMUST00000148193.2
|
Cfap69
|
cilia and flagella associated protein 69 |
| chr15_-_97808502 | 0.01 |
ENSMUST00000173104.8
ENSMUST00000174633.8 |
Vdr
|
vitamin D (1,25-dihydroxyvitamin D3) receptor |
| chr14_+_53220658 | 0.00 |
ENSMUST00000200548.2
|
Trav9d-4
|
T cell receptor alpha variable 9D-4 |
| chr18_+_35731735 | 0.00 |
ENSMUST00000236868.2
|
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0097037 | heme export(GO:0097037) |
| 2.7 | 8.1 | GO:0034378 | chylomicron assembly(GO:0034378) regulation of chylomicron remnant clearance(GO:0090320) |
| 2.7 | 8.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 2.1 | 8.6 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 1.3 | 4.0 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.3 | 9.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.1 | 4.5 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.7 | 2.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.4 | 1.9 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.3 | 1.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.3 | 8.8 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.3 | 5.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.3 | 2.6 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 1.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 3.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 4.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 1.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 1.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 2.7 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.7 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 1.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 1.3 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 3.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 9.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.5 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.1 | 0.8 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 6.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.3 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 2.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.5 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 2.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 1.8 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.0 | 7.6 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 20.2 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.8 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.0 | 5.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 5.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 2.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.2 | 3.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 9.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 8.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 5.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 2.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 2.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 5.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 6.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 13.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 4.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 2.7 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 6.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 6.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 27.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 3.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 2.7 | 8.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.8 | 8.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 1.5 | 9.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.4 | 5.6 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.8 | 5.9 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.5 | 1.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 6.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.5 | 2.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.5 | 9.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.4 | 4.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 6.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.3 | 2.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.3 | 1.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 1.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 25.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 3.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 19.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 5.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 3.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 2.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.6 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 8.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 5.7 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 8.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 4.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 30.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 28.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 3.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 6.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 18.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 9.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 6.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 4.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |