Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
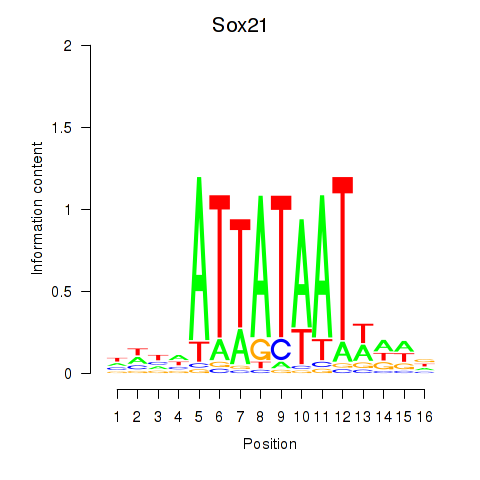
Results for Sox21
Z-value: 0.51
Transcription factors associated with Sox21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox21
|
ENSMUSG00000061517.9 | SRY (sex determining region Y)-box 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox21 | mm39_v1_chr14_-_118474404_118474448 | -0.01 | 9.4e-01 | Click! |
Activity profile of Sox21 motif
Sorted Z-values of Sox21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_115420876 | 2.66 |
ENSMUST00000126645.8
ENSMUST00000030480.4 |
Cyp4a31
|
cytochrome P450, family 4, subfamily a, polypeptide 31 |
| chr6_-_41012435 | 2.59 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr19_-_39875192 | 2.13 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr16_+_36097313 | 1.86 |
ENSMUST00000232150.2
|
Stfa1
|
stefin A1 |
| chr16_+_36097505 | 1.44 |
ENSMUST00000042097.11
|
Stfa1
|
stefin A1 |
| chr16_-_18904240 | 1.42 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr7_-_142253247 | 1.31 |
ENSMUST00000105934.8
|
Ins2
|
insulin II |
| chr3_-_10273628 | 1.25 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr5_-_87716882 | 1.23 |
ENSMUST00000113314.3
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr6_+_67586695 | 1.18 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr2_+_174292471 | 1.13 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr7_+_30193047 | 1.10 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr14_-_56322654 | 0.97 |
ENSMUST00000015594.9
|
Mcpt8
|
mast cell protease 8 |
| chr9_+_7558449 | 0.85 |
ENSMUST00000018765.4
|
Mmp8
|
matrix metallopeptidase 8 |
| chr13_-_93774469 | 0.84 |
ENSMUST00000099309.6
|
Bhmt
|
betaine-homocysteine methyltransferase |
| chr2_-_28511941 | 0.74 |
ENSMUST00000028156.8
ENSMUST00000164290.8 |
Gfi1b
|
growth factor independent 1B |
| chr17_-_32639936 | 0.71 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr6_-_78355834 | 0.71 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr18_-_15536747 | 0.69 |
ENSMUST00000079081.8
|
Aqp4
|
aquaporin 4 |
| chr6_+_68026941 | 0.66 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr3_-_75177378 | 0.63 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr15_-_60696790 | 0.61 |
ENSMUST00000100635.5
|
Lratd2
|
LRAT domain containing 1 |
| chr13_-_33035150 | 0.54 |
ENSMUST00000091668.13
ENSMUST00000076352.8 |
Serpinb1a
|
serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| chr3_-_82957104 | 0.52 |
ENSMUST00000048246.5
|
Fgb
|
fibrinogen beta chain |
| chr16_-_22848153 | 0.49 |
ENSMUST00000232459.2
|
Kng2
|
kininogen 2 |
| chr17_-_71305003 | 0.48 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_-_30988381 | 0.47 |
ENSMUST00000232841.2
|
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr11_+_69855584 | 0.46 |
ENSMUST00000108597.8
ENSMUST00000060651.6 ENSMUST00000108596.8 |
Cldn7
|
claudin 7 |
| chr13_+_93810911 | 0.45 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr12_-_114398864 | 0.44 |
ENSMUST00000103489.2
|
Ighv6-6
|
immunoglobulin heavy variable 6-6 |
| chr12_-_102390000 | 0.41 |
ENSMUST00000110020.8
|
Lgmn
|
legumain |
| chr6_-_16898440 | 0.41 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr9_+_103940575 | 0.40 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr9_+_103940879 | 0.40 |
ENSMUST00000047799.13
ENSMUST00000189998.3 |
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr7_+_27977141 | 0.36 |
ENSMUST00000094651.4
|
Eid2b
|
EP300 interacting inhibitor of differentiation 2B |
| chr6_+_41515152 | 0.35 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chrM_+_11735 | 0.35 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr13_-_19803928 | 0.34 |
ENSMUST00000221014.2
ENSMUST00000002885.8 ENSMUST00000220944.2 |
Epdr1
|
ependymin related protein 1 (zebrafish) |
| chr9_+_108880221 | 0.32 |
ENSMUST00000200629.5
ENSMUST00000200515.5 ENSMUST00000197689.5 ENSMUST00000196954.5 ENSMUST00000198376.5 ENSMUST00000197483.5 ENSMUST00000198295.5 |
Shisa5
|
shisa family member 5 |
| chr17_+_48047955 | 0.31 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr1_+_62742444 | 0.31 |
ENSMUST00000102822.9
ENSMUST00000075144.12 |
Nrp2
|
neuropilin 2 |
| chr19_-_8775817 | 0.30 |
ENSMUST00000235964.2
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr11_+_87590720 | 0.28 |
ENSMUST00000040089.5
|
Rnf43
|
ring finger protein 43 |
| chr19_-_8775935 | 0.27 |
ENSMUST00000096261.5
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr8_-_41507808 | 0.27 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr9_+_108880158 | 0.25 |
ENSMUST00000198708.5
|
Shisa5
|
shisa family member 5 |
| chr19_+_11495858 | 0.25 |
ENSMUST00000025580.10
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr3_-_59127571 | 0.23 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr11_+_101221431 | 0.22 |
ENSMUST00000103105.10
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr11_+_114566257 | 0.22 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr10_-_129738595 | 0.22 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr3_-_130503041 | 0.21 |
ENSMUST00000043937.9
|
Ostc
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr6_-_69877961 | 0.21 |
ENSMUST00000197290.2
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr2_-_35869636 | 0.21 |
ENSMUST00000028248.11
ENSMUST00000112976.9 |
Ttll11
|
tubulin tyrosine ligase-like family, member 11 |
| chr15_+_31602252 | 0.21 |
ENSMUST00000042702.7
ENSMUST00000161061.3 |
Atpsckmt
|
ATP synthase C subunit lysine N-methyltransferase |
| chr10_-_62178453 | 0.20 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr1_+_107439145 | 0.20 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr12_+_103524690 | 0.19 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr3_-_16060545 | 0.19 |
ENSMUST00000194367.6
|
Gm5150
|
predicted gene 5150 |
| chr5_+_146769700 | 0.19 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chrX_+_164953444 | 0.18 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr15_+_102378966 | 0.18 |
ENSMUST00000077037.13
ENSMUST00000229102.2 ENSMUST00000229618.2 ENSMUST00000229275.2 ENSMUST00000231089.2 ENSMUST00000229802.2 ENSMUST00000229854.2 ENSMUST00000108838.5 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr7_+_80511856 | 0.18 |
ENSMUST00000044115.9
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr17_+_6697511 | 0.17 |
ENSMUST00000179569.3
|
Dynlt1b
|
dynein light chain Tctex-type 1B |
| chr15_-_6904450 | 0.16 |
ENSMUST00000022746.13
ENSMUST00000176826.2 |
Osmr
|
oncostatin M receptor |
| chr3_-_14843512 | 0.16 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chrX_-_56371953 | 0.16 |
ENSMUST00000176986.8
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chrM_-_14061 | 0.15 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr10_+_42736771 | 0.15 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chrM_+_10167 | 0.15 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_+_138047536 | 0.14 |
ENSMUST00000199673.6
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr10_+_23952398 | 0.13 |
ENSMUST00000051133.6
|
Taar8a
|
trace amine-associated receptor 8A |
| chr1_-_139786421 | 0.13 |
ENSMUST00000194186.6
ENSMUST00000094489.5 ENSMUST00000239380.2 |
Cfhr2
|
complement factor H-related 2 |
| chr17_+_6869070 | 0.13 |
ENSMUST00000092966.5
|
Dynlt1c
|
dynein light chain Tctex-type 1C |
| chr10_-_13264497 | 0.12 |
ENSMUST00000105546.8
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr18_+_37637317 | 0.12 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr18_+_37827413 | 0.11 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr6_-_58418303 | 0.11 |
ENSMUST00000228577.2
ENSMUST00000227466.2 |
Vmn1r30
|
vomeronasal 1 receptor 30 |
| chr8_-_18791557 | 0.10 |
ENSMUST00000033846.7
|
Angpt2
|
angiopoietin 2 |
| chr17_-_6367692 | 0.10 |
ENSMUST00000232499.2
ENSMUST00000169415.3 |
Dynlt1a
|
dynein light chain Tctex-type 1A |
| chr10_+_127685785 | 0.10 |
ENSMUST00000077530.3
|
Rdh19
|
retinol dehydrogenase 19 |
| chr17_+_29487881 | 0.09 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr2_-_25517945 | 0.09 |
ENSMUST00000028307.9
|
Fcna
|
ficolin A |
| chr2_-_140513382 | 0.09 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr3_+_94350622 | 0.08 |
ENSMUST00000029786.14
ENSMUST00000196143.2 |
Mrpl9
|
mitochondrial ribosomal protein L9 |
| chr2_-_155424576 | 0.08 |
ENSMUST00000126322.8
|
Gss
|
glutathione synthetase |
| chr6_-_99412306 | 0.08 |
ENSMUST00000113322.9
ENSMUST00000176850.8 ENSMUST00000176632.8 |
Foxp1
|
forkhead box P1 |
| chr17_+_69569184 | 0.08 |
ENSMUST00000224951.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr8_-_35432783 | 0.08 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr1_-_171854818 | 0.07 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr2_-_87868043 | 0.07 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73 |
| chr19_+_7534816 | 0.07 |
ENSMUST00000136465.8
ENSMUST00000025925.11 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr4_+_102278715 | 0.07 |
ENSMUST00000106904.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr8_+_94537910 | 0.07 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr17_+_80614795 | 0.06 |
ENSMUST00000223878.2
ENSMUST00000068175.6 ENSMUST00000224391.2 |
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr10_-_23977810 | 0.06 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr10_-_28862289 | 0.06 |
ENSMUST00000152363.8
ENSMUST00000015663.7 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr7_+_128289261 | 0.06 |
ENSMUST00000151237.5
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr18_+_37822865 | 0.06 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2 |
| chr10_+_129306867 | 0.05 |
ENSMUST00000213222.2
|
Olfr788
|
olfactory receptor 788 |
| chr17_+_35561218 | 0.05 |
ENSMUST00000074806.12
|
H2-Q2
|
histocompatibility 2, Q region locus 2 |
| chr3_-_19319155 | 0.05 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr11_+_78717398 | 0.05 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr11_+_22462088 | 0.05 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr15_-_99717956 | 0.05 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr10_-_38998272 | 0.05 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr5_+_143389573 | 0.04 |
ENSMUST00000110731.4
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr18_-_67682312 | 0.04 |
ENSMUST00000224799.2
|
Spire1
|
spire type actin nucleation factor 1 |
| chr19_+_7534838 | 0.04 |
ENSMUST00000141887.8
ENSMUST00000136756.2 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr12_-_115083839 | 0.04 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr3_+_32490525 | 0.04 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr11_+_119283887 | 0.03 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr2_+_87696836 | 0.03 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr10_-_10348058 | 0.03 |
ENSMUST00000179956.8
ENSMUST00000208717.2 ENSMUST00000172530.8 ENSMUST00000132573.2 |
Adgb
|
androglobin |
| chr1_-_4563821 | 0.03 |
ENSMUST00000191939.2
|
Sox17
|
SRY (sex determining region Y)-box 17 |
| chr12_-_115567853 | 0.03 |
ENSMUST00000103538.3
ENSMUST00000198646.2 |
Ighv1-67
|
immunoglobulin heavy variable V1-67 |
| chr3_+_105778174 | 0.02 |
ENSMUST00000164730.2
ENSMUST00000010279.10 |
Adora3
Tmigd3
|
adenosine A3 receptor transmembrane and immunoglobulin domain containing 3 |
| chr17_+_29487762 | 0.02 |
ENSMUST00000064709.13
ENSMUST00000234711.2 |
BC004004
|
cDNA sequence BC004004 |
| chr5_+_20112704 | 0.02 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_+_83985495 | 0.02 |
ENSMUST00000113821.8
|
Dysf
|
dysferlin |
| chr7_-_121580930 | 0.02 |
ENSMUST00000205438.2
ENSMUST00000057576.8 |
Cog7
|
component of oligomeric golgi complex 7 |
| chr16_-_26786995 | 0.02 |
ENSMUST00000231969.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr4_-_52859227 | 0.01 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273 |
| chr2_+_88470886 | 0.01 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr3_+_19562753 | 0.01 |
ENSMUST00000118968.8
|
Dnajc5b
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chrX_+_13499008 | 0.01 |
ENSMUST00000096492.4
|
Gpr34
|
G protein-coupled receptor 34 |
| chr2_-_79738773 | 0.01 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_-_104250951 | 0.01 |
ENSMUST00000216750.2
ENSMUST00000215538.2 |
Olfr655
|
olfactory receptor 655 |
| chr1_-_75200298 | 0.01 |
ENSMUST00000179573.2
|
A630095N17Rik
|
RIKEN cDNA A630095N17 gene |
| chr9_+_103917821 | 0.01 |
ENSMUST00000216593.2
ENSMUST00000147249.3 |
Nphp3
Gm28305
|
nephronophthisis 3 (adolescent) predicted gene 28305 |
| chr14_+_26414422 | 0.00 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr9_-_39237341 | 0.00 |
ENSMUST00000216132.2
|
Olfr948
|
olfactory receptor 948 |
| chr2_+_67004178 | 0.00 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox21
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.4 | 1.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.8 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.1 | 0.7 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.7 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.7 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 | 0.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 1.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.2 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.2 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.6 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 1.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 3.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.0 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 1.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 2.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.0 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 1.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0050051 | alkane 1-monooxygenase activity(GO:0018685) leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.3 | 1.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 0.8 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.7 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.2 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 5.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |