Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
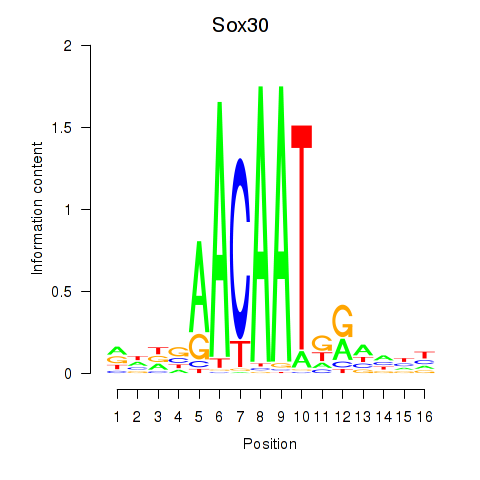
Results for Sox30
Z-value: 0.58
Transcription factors associated with Sox30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox30
|
ENSMUSG00000040489.6 | SRY (sex determining region Y)-box 30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox30 | mm39_v1_chr11_+_45871135_45871164 | 0.18 | 3.0e-01 | Click! |
Activity profile of Sox30 motif
Sorted Z-values of Sox30 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_113198765 | 5.08 |
ENSMUST00000179568.3
|
Amy2a4
|
amylase 2a4 |
| chr17_-_31363245 | 3.22 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr6_-_136899167 | 2.72 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr19_+_44282113 | 2.68 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr7_+_43874752 | 2.15 |
ENSMUST00000075162.5
|
Klk1
|
kallikrein 1 |
| chr4_-_119047202 | 2.03 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047167 | 1.68 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chrX_+_92698469 | 1.67 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr4_-_119047146 | 1.31 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047180 | 1.28 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr1_+_135656885 | 1.21 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr5_+_146769700 | 1.14 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chr6_-_136758716 | 1.11 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr9_-_103357564 | 1.07 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr10_-_62628008 | 1.04 |
ENSMUST00000217768.2
ENSMUST00000020268.7 ENSMUST00000218946.2 ENSMUST00000219527.2 |
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr14_+_117162791 | 0.96 |
ENSMUST00000088483.10
|
Gpc6
|
glypican 6 |
| chr3_-_102871440 | 0.95 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr17_-_79662514 | 0.84 |
ENSMUST00000068958.9
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr5_-_137101108 | 0.81 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr19_+_34194990 | 0.77 |
ENSMUST00000119603.2
|
Stambpl1
|
STAM binding protein like 1 |
| chr2_-_84652890 | 0.66 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr4_+_109200225 | 0.63 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr6_-_87312743 | 0.61 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr3_+_90383425 | 0.59 |
ENSMUST00000001042.10
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr11_+_3933699 | 0.55 |
ENSMUST00000063004.14
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr10_+_96453408 | 0.52 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr11_+_3933636 | 0.51 |
ENSMUST00000078757.8
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr4_-_116263183 | 0.48 |
ENSMUST00000123072.8
ENSMUST00000144281.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr19_-_24454720 | 0.45 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr11_-_99134885 | 0.42 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr1_-_176641607 | 0.37 |
ENSMUST00000195717.6
ENSMUST00000192961.6 |
Cep170
|
centrosomal protein 170 |
| chr10_+_17672004 | 0.37 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chr18_-_68562385 | 0.36 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr10_-_37014859 | 0.34 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr9_-_72019053 | 0.34 |
ENSMUST00000183492.8
ENSMUST00000184523.8 ENSMUST00000034755.13 |
Tcf12
|
transcription factor 12 |
| chr9_-_72019109 | 0.34 |
ENSMUST00000183404.8
ENSMUST00000184783.8 |
Tcf12
|
transcription factor 12 |
| chr6_-_24168082 | 0.31 |
ENSMUST00000031713.9
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr1_+_38037086 | 0.30 |
ENSMUST00000027252.8
|
Eif5b
|
eukaryotic translation initiation factor 5B |
| chr3_-_9029097 | 0.30 |
ENSMUST00000091354.12
ENSMUST00000094381.11 |
Tpd52
|
tumor protein D52 |
| chr3_-_9898713 | 0.27 |
ENSMUST00000161949.8
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr9_-_72018453 | 0.25 |
ENSMUST00000183992.8
|
Tcf12
|
transcription factor 12 |
| chr5_+_139197689 | 0.24 |
ENSMUST00000148772.8
ENSMUST00000110882.8 |
Sun1
|
Sad1 and UNC84 domain containing 1 |
| chr13_-_43632368 | 0.24 |
ENSMUST00000222651.2
|
Ranbp9
|
RAN binding protein 9 |
| chr1_-_151376544 | 0.23 |
ENSMUST00000187991.2
ENSMUST00000187048.7 ENSMUST00000186415.7 |
Rnf2
|
ring finger protein 2 |
| chr9_+_7764042 | 0.23 |
ENSMUST00000052865.16
|
Tmem123
|
transmembrane protein 123 |
| chr12_-_99849660 | 0.23 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr11_+_21041291 | 0.22 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr2_+_3115250 | 0.22 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr8_-_86281946 | 0.21 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr2_+_127750978 | 0.20 |
ENSMUST00000110344.2
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr18_-_88945571 | 0.18 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr19_-_56810593 | 0.17 |
ENSMUST00000118592.8
|
Ccdc186
|
coiled-coil domain containing 186 |
| chr9_+_39985125 | 0.16 |
ENSMUST00000054051.5
|
Olfr982
|
olfactory receptor 982 |
| chr6_-_12749192 | 0.16 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr9_+_111014254 | 0.16 |
ENSMUST00000198986.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr11_-_106606076 | 0.15 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr9_-_60418286 | 0.15 |
ENSMUST00000098660.10
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr17_-_34337446 | 0.12 |
ENSMUST00000095347.13
|
Brd2
|
bromodomain containing 2 |
| chr9_-_19968858 | 0.12 |
ENSMUST00000216538.2
ENSMUST00000212098.3 |
Olfr867
|
olfactory receptor 867 |
| chr11_-_106605772 | 0.12 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr9_+_7571397 | 0.11 |
ENSMUST00000120900.8
ENSMUST00000151853.8 ENSMUST00000152878.3 |
Mmp27
|
matrix metallopeptidase 27 |
| chr5_-_136275407 | 0.11 |
ENSMUST00000196245.2
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr6_-_129449739 | 0.10 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr9_-_72018933 | 0.09 |
ENSMUST00000185117.8
|
Tcf12
|
transcription factor 12 |
| chr4_+_109092610 | 0.09 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr4_+_136011969 | 0.09 |
ENSMUST00000144217.8
|
Zfp46
|
zinc finger protein 46 |
| chr3_-_9898676 | 0.08 |
ENSMUST00000108384.9
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr11_-_58521327 | 0.07 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr2_-_119617985 | 0.06 |
ENSMUST00000110793.8
ENSMUST00000099529.9 ENSMUST00000048493.12 |
Rpap1
|
RNA polymerase II associated protein 1 |
| chr4_+_109092459 | 0.06 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr18_+_37875135 | 0.06 |
ENSMUST00000003599.9
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr2_-_181240921 | 0.06 |
ENSMUST00000060173.9
|
Samd10
|
sterile alpha motif domain containing 10 |
| chr17_+_53786240 | 0.06 |
ENSMUST00000017975.7
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr14_-_19873839 | 0.06 |
ENSMUST00000022341.7
|
Rtraf
|
RNA transcription, translation and transport factor |
| chr11_-_116572116 | 0.06 |
ENSMUST00000144398.2
|
St6galnac2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr18_+_37858753 | 0.05 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr18_+_37130860 | 0.04 |
ENSMUST00000115659.6
|
Pcdha9
|
protocadherin alpha 9 |
| chr11_+_51152546 | 0.04 |
ENSMUST00000130641.8
|
Clk4
|
CDC like kinase 4 |
| chr4_+_83335947 | 0.04 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr7_+_43661592 | 0.03 |
ENSMUST00000048945.6
|
Klk1b26
|
kallikrein 1-related petidase b26 |
| chr5_+_102992873 | 0.03 |
ENSMUST00000070000.6
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr6_-_12749409 | 0.03 |
ENSMUST00000119581.7
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr4_-_73869071 | 0.02 |
ENSMUST00000095023.2
ENSMUST00000030101.4 |
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene |
| chr18_+_37610858 | 0.02 |
ENSMUST00000051442.7
|
Pcdhb16
|
protocadherin beta 16 |
| chr2_+_85545763 | 0.02 |
ENSMUST00000216443.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr11_+_67128843 | 0.01 |
ENSMUST00000018632.11
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle |
| chr4_-_114991478 | 0.01 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox30
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.4 | 2.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 0.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 1.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.3 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.1 | 1.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.6 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0043578 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.6 | GO:1905050 | positive regulation of metallopeptidase activity(GO:1905050) |
| 0.0 | 0.5 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 1.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 1.7 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.4 | 2.7 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.4 | 1.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 1.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.2 | 3.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 2.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |