Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
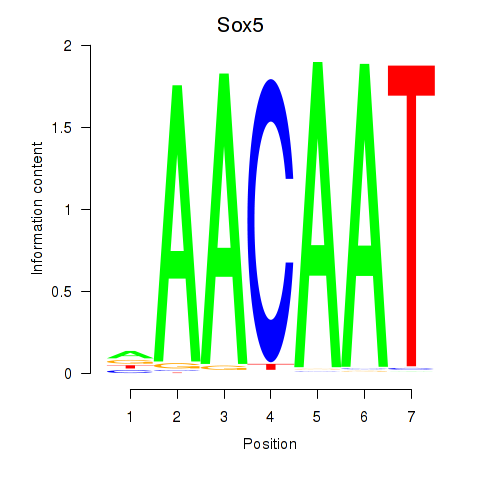
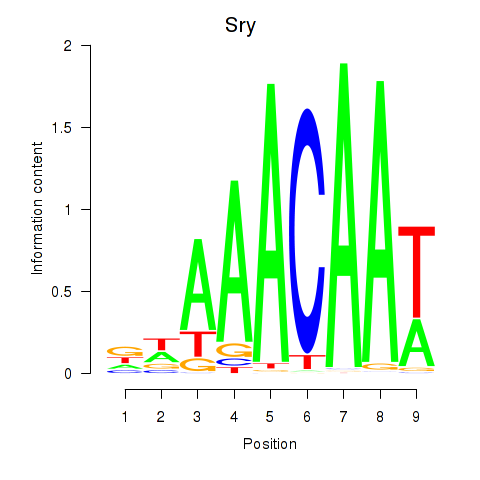
Results for Sox5_Sry
Z-value: 1.50
Transcription factors associated with Sox5_Sry
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox5
|
ENSMUSG00000041540.17 | SRY (sex determining region Y)-box 5 |
|
Sry
|
ENSMUSG00000069036.4 | sex determining region of Chr Y |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox5 | mm39_v1_chr6_-_144155197_144155294 | 0.47 | 3.9e-03 | Click! |
Activity profile of Sox5_Sry motif
Sorted Z-values of Sox5_Sry motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_87695352 | 11.64 |
ENSMUST00000121952.8
ENSMUST00000126490.8 |
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87695117 | 9.84 |
ENSMUST00000105300.9
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_87694924 | 8.35 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr6_+_17491231 | 6.26 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr10_+_87697155 | 6.14 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr2_+_102488985 | 5.03 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr14_-_52151537 | 5.01 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr12_+_40495951 | 4.59 |
ENSMUST00000037488.8
|
Dock4
|
dedicator of cytokinesis 4 |
| chr12_-_83643964 | 4.51 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr14_-_100522101 | 4.45 |
ENSMUST00000228216.2
|
Klf12
|
Kruppel-like factor 12 |
| chr19_-_40175709 | 4.43 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr12_-_83643883 | 4.34 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr8_-_95405234 | 4.17 |
ENSMUST00000213043.2
|
Pllp
|
plasma membrane proteolipid |
| chr4_+_129229373 | 4.04 |
ENSMUST00000141235.8
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr9_+_47441471 | 3.70 |
ENSMUST00000114548.8
ENSMUST00000152459.8 ENSMUST00000143026.9 ENSMUST00000085909.9 ENSMUST00000114547.8 ENSMUST00000239368.2 ENSMUST00000214542.2 ENSMUST00000034581.4 |
Cadm1
|
cell adhesion molecule 1 |
| chr5_-_66238313 | 3.69 |
ENSMUST00000202700.4
ENSMUST00000094757.9 ENSMUST00000113724.6 |
Rbm47
|
RNA binding motif protein 47 |
| chr8_-_85500010 | 3.52 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr1_-_72251466 | 3.41 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr12_-_99359265 | 3.26 |
ENSMUST00000177451.8
|
Foxn3
|
forkhead box N3 |
| chr18_-_60881679 | 3.24 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr2_-_6726998 | 3.16 |
ENSMUST00000182657.2
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr6_-_71417607 | 2.95 |
ENSMUST00000002292.15
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr19_+_23118545 | 2.90 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9 |
| chr11_+_93935021 | 2.89 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr15_-_96917804 | 2.87 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr14_-_100521888 | 2.83 |
ENSMUST00000226774.2
|
Klf12
|
Kruppel-like factor 12 |
| chr5_-_92231517 | 2.76 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr5_+_42225303 | 2.73 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr4_+_140970161 | 2.65 |
ENSMUST00000138096.8
ENSMUST00000006618.9 ENSMUST00000125392.8 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr4_+_144619647 | 2.62 |
ENSMUST00000154208.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_144619696 | 2.61 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr11_+_93935156 | 2.58 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr5_-_87572060 | 2.51 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr11_+_93935066 | 2.42 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr4_-_82423511 | 2.42 |
ENSMUST00000050872.15
ENSMUST00000064770.9 |
Nfib
|
nuclear factor I/B |
| chr2_-_62242562 | 2.41 |
ENSMUST00000047812.8
|
Dpp4
|
dipeptidylpeptidase 4 |
| chr12_+_100745333 | 2.31 |
ENSMUST00000110073.8
ENSMUST00000110069.8 |
Dglucy
|
D-glutamate cyclase |
| chr10_+_116111441 | 2.31 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr9_+_74883377 | 2.30 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr8_-_85500998 | 2.30 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr19_-_4384029 | 2.29 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr6_-_93889483 | 2.27 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr6_+_54249817 | 2.26 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr18_-_3337467 | 2.18 |
ENSMUST00000154135.8
ENSMUST00000142690.2 ENSMUST00000025069.11 ENSMUST00000165086.8 ENSMUST00000082141.12 ENSMUST00000149803.8 |
Crem
|
cAMP responsive element modulator |
| chr18_-_16942289 | 2.17 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr15_-_85918378 | 2.15 |
ENSMUST00000016172.10
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr7_+_119499322 | 2.13 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr17_+_44445659 | 2.08 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr2_-_6726701 | 2.06 |
ENSMUST00000114927.9
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr16_-_28748410 | 2.04 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr12_+_100745314 | 2.04 |
ENSMUST00000069782.11
|
Dglucy
|
D-glutamate cyclase |
| chr11_-_86884507 | 2.03 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr10_+_116137277 | 2.03 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr9_+_70114623 | 2.02 |
ENSMUST00000034745.9
|
Myo1e
|
myosin IE |
| chr9_-_35028100 | 2.02 |
ENSMUST00000034537.8
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr9_-_29323032 | 2.01 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr5_-_92231314 | 1.99 |
ENSMUST00000169094.8
ENSMUST00000167918.8 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr7_+_16044336 | 1.96 |
ENSMUST00000136781.2
|
Bbc3
|
BCL2 binding component 3 |
| chr4_+_43641262 | 1.93 |
ENSMUST00000123351.8
ENSMUST00000128549.3 |
Npr2
|
natriuretic peptide receptor 2 |
| chr1_-_133849131 | 1.92 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr2_-_104324035 | 1.92 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr2_+_125876883 | 1.90 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr6_-_116084810 | 1.90 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr16_+_19916292 | 1.83 |
ENSMUST00000023509.5
ENSMUST00000232088.2 ENSMUST00000231842.2 |
Klhl24
|
kelch-like 24 |
| chr7_+_16043502 | 1.81 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr1_+_16175998 | 1.81 |
ENSMUST00000027053.8
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr17_-_46956920 | 1.79 |
ENSMUST00000233974.2
|
Klc4
|
kinesin light chain 4 |
| chrM_-_14061 | 1.78 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr6_+_91661074 | 1.74 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr8_+_57004125 | 1.71 |
ENSMUST00000110322.9
ENSMUST00000040218.13 ENSMUST00000210863.2 |
Fbxo8
|
F-box protein 8 |
| chr4_-_109013807 | 1.71 |
ENSMUST00000161363.2
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr6_-_93769426 | 1.70 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr6_+_91661034 | 1.69 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr15_+_25622611 | 1.68 |
ENSMUST00000110457.8
ENSMUST00000137601.8 |
Myo10
|
myosin X |
| chr18_-_84104507 | 1.67 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr9_+_119978773 | 1.67 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr10_-_18110682 | 1.66 |
ENSMUST00000052648.9
ENSMUST00000080860.13 ENSMUST00000173243.8 |
Ccdc28a
|
coiled-coil domain containing 28A |
| chr10_+_34359395 | 1.64 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr1_+_143516402 | 1.61 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr11_+_97554192 | 1.61 |
ENSMUST00000044730.12
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
| chr4_-_82623972 | 1.60 |
ENSMUST00000155821.2
|
Nfib
|
nuclear factor I/B |
| chr9_-_29323500 | 1.58 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr1_+_176642226 | 1.58 |
ENSMUST00000056773.15
ENSMUST00000027785.15 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr4_-_82423985 | 1.57 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr8_-_45835234 | 1.56 |
ENSMUST00000093526.13
|
Fam149a
|
family with sequence similarity 149, member A |
| chr4_+_122889407 | 1.55 |
ENSMUST00000144998.2
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr18_-_84104574 | 1.55 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr6_-_71609881 | 1.53 |
ENSMUST00000065509.11
ENSMUST00000207023.2 |
Kdm3a
|
lysine (K)-specific demethylase 3A |
| chr10_-_68114543 | 1.51 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr5_+_137568982 | 1.51 |
ENSMUST00000196471.5
ENSMUST00000198783.5 |
Tfr2
|
transferrin receptor 2 |
| chr11_-_60243695 | 1.50 |
ENSMUST00000095254.12
ENSMUST00000102683.11 ENSMUST00000093048.13 ENSMUST00000093046.13 ENSMUST00000064019.15 ENSMUST00000102682.5 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr3_+_85878376 | 1.49 |
ENSMUST00000238443.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr4_-_45489794 | 1.48 |
ENSMUST00000146236.8
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr13_+_46822992 | 1.48 |
ENSMUST00000099547.4
|
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr6_-_47790272 | 1.48 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr2_-_37593287 | 1.47 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr2_+_6327431 | 1.46 |
ENSMUST00000114937.8
|
Usp6nl
|
USP6 N-terminal like |
| chr18_+_67338437 | 1.46 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr16_+_34815177 | 1.45 |
ENSMUST00000231589.2
|
Mylk
|
myosin, light polypeptide kinase |
| chr5_-_34345014 | 1.45 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr3_+_135531548 | 1.45 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr4_-_82423944 | 1.44 |
ENSMUST00000107248.8
ENSMUST00000107247.8 |
Nfib
|
nuclear factor I/B |
| chr3_+_13536696 | 1.43 |
ENSMUST00000191806.3
ENSMUST00000193117.3 |
Ralyl
|
RALY RNA binding protein-like |
| chr11_-_98220466 | 1.40 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr18_-_3337679 | 1.38 |
ENSMUST00000150235.8
ENSMUST00000154470.8 |
Crem
|
cAMP responsive element modulator |
| chr5_+_30869193 | 1.37 |
ENSMUST00000088081.11
ENSMUST00000101442.4 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr2_+_27567246 | 1.35 |
ENSMUST00000166775.8
|
Rxra
|
retinoid X receptor alpha |
| chr4_-_103072343 | 1.32 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr4_-_33248499 | 1.32 |
ENSMUST00000049357.10
|
Pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr3_-_91990439 | 1.30 |
ENSMUST00000058150.8
|
Lor
|
loricrin |
| chr2_+_27567213 | 1.28 |
ENSMUST00000077257.12
|
Rxra
|
retinoid X receptor alpha |
| chr10_+_34359513 | 1.28 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr2_+_125876566 | 1.25 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr14_-_47805861 | 1.24 |
ENSMUST00000228784.2
ENSMUST00000042988.7 |
Atg14
|
autophagy related 14 |
| chr14_-_33169099 | 1.24 |
ENSMUST00000111944.10
ENSMUST00000022504.12 ENSMUST00000111945.9 |
Mapk8
|
mitogen-activated protein kinase 8 |
| chr3_+_95801263 | 1.24 |
ENSMUST00000015894.12
|
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chrX_+_139808351 | 1.24 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr13_-_111626562 | 1.23 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr8_-_70355208 | 1.20 |
ENSMUST00000110167.5
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr16_+_78098344 | 1.20 |
ENSMUST00000232148.2
ENSMUST00000023572.15 |
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr2_+_4722956 | 1.20 |
ENSMUST00000056914.7
|
Bend7
|
BEN domain containing 7 |
| chr9_-_50639230 | 1.19 |
ENSMUST00000118707.2
ENSMUST00000034566.15 |
Dixdc1
|
DIX domain containing 1 |
| chr12_-_40495753 | 1.19 |
ENSMUST00000069692.10
ENSMUST00000069637.15 |
Zfp277
|
zinc finger protein 277 |
| chr3_+_63203235 | 1.19 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr5_+_30971915 | 1.18 |
ENSMUST00000031058.15
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr11_-_110142565 | 1.18 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr5_+_30868908 | 1.17 |
ENSMUST00000114729.8
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr10_+_69542153 | 1.16 |
ENSMUST00000182992.8
|
Ank3
|
ankyrin 3, epithelial |
| chr11_+_93934940 | 1.15 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr1_-_130867810 | 1.15 |
ENSMUST00000112465.2
ENSMUST00000187410.7 ENSMUST00000187916.7 |
Il19
|
interleukin 19 |
| chr4_+_11579648 | 1.15 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chrX_+_71025128 | 1.15 |
ENSMUST00000114569.2
|
Fate1
|
fetal and adult testis expressed 1 |
| chr1_+_131898325 | 1.14 |
ENSMUST00000027695.8
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr6_-_114898739 | 1.14 |
ENSMUST00000032459.14
|
Vgll4
|
vestigial like family member 4 |
| chr13_+_83720484 | 1.13 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr13_+_41040657 | 1.12 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr6_+_34686543 | 1.12 |
ENSMUST00000031775.13
|
Cald1
|
caldesmon 1 |
| chrX_+_72800675 | 1.12 |
ENSMUST00000002079.7
|
Plxnb3
|
plexin B3 |
| chr16_+_42727926 | 1.12 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_+_128290204 | 1.12 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr19_-_41195213 | 1.11 |
ENSMUST00000169941.2
ENSMUST00000025986.15 |
Tll2
|
tolloid-like 2 |
| chr3_+_63202940 | 1.10 |
ENSMUST00000194150.6
|
Mme
|
membrane metallo endopeptidase |
| chr7_-_119494669 | 1.10 |
ENSMUST00000098080.9
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr6_+_134012602 | 1.10 |
ENSMUST00000081028.13
ENSMUST00000111963.8 |
Etv6
|
ets variant 6 |
| chr5_+_30972067 | 1.10 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr16_-_78373510 | 1.07 |
ENSMUST00000231973.2
ENSMUST00000232528.2 ENSMUST00000114220.9 |
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr15_+_102011352 | 1.07 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr15_+_102011415 | 1.06 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr16_-_78373537 | 1.05 |
ENSMUST00000232052.2
ENSMUST00000114219.8 ENSMUST00000114218.2 |
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr15_+_34082805 | 1.05 |
ENSMUST00000022865.17
|
Mtdh
|
metadherin |
| chr10_+_57362512 | 1.05 |
ENSMUST00000220042.2
|
Hsf2
|
heat shock factor 2 |
| chr18_+_69477541 | 1.02 |
ENSMUST00000114985.10
ENSMUST00000128706.8 ENSMUST00000201781.4 ENSMUST00000202674.4 |
Tcf4
|
transcription factor 4 |
| chr14_-_48900192 | 1.02 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chrX_+_99811325 | 1.02 |
ENSMUST00000000901.13
ENSMUST00000113736.9 ENSMUST00000087984.11 |
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr16_-_95260104 | 1.02 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr9_+_7764042 | 1.01 |
ENSMUST00000052865.16
|
Tmem123
|
transmembrane protein 123 |
| chr4_+_109835224 | 1.00 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr1_+_132996237 | 0.99 |
ENSMUST00000239467.2
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr11_+_72498029 | 0.98 |
ENSMUST00000021148.13
ENSMUST00000138247.8 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr11_+_118913788 | 0.97 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr9_+_117869543 | 0.97 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr3_+_90161470 | 0.96 |
ENSMUST00000029545.15
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr4_-_70453140 | 0.96 |
ENSMUST00000107359.9
|
Megf9
|
multiple EGF-like-domains 9 |
| chr2_-_6327884 | 0.96 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene |
| chr19_-_8196196 | 0.95 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr4_+_122889737 | 0.93 |
ENSMUST00000106252.9
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
| chr5_+_122239030 | 0.92 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr6_+_120341055 | 0.92 |
ENSMUST00000005108.10
|
Kdm5a
|
lysine (K)-specific demethylase 5A |
| chr14_-_55231998 | 0.92 |
ENSMUST00000227518.2
ENSMUST00000226424.2 ENSMUST00000153783.2 ENSMUST00000102803.11 ENSMUST00000168485.8 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr10_+_57362457 | 0.92 |
ENSMUST00000079833.6
|
Hsf2
|
heat shock factor 2 |
| chr6_+_34686373 | 0.91 |
ENSMUST00000115021.8
|
Cald1
|
caldesmon 1 |
| chr1_+_75526225 | 0.91 |
ENSMUST00000154101.8
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr5_+_65288418 | 0.90 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr6_+_15185202 | 0.90 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr5_+_122239007 | 0.89 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr1_+_153776323 | 0.89 |
ENSMUST00000140685.4
ENSMUST00000139476.8 |
Glul
|
glutamate-ammonia ligase (glutamine synthetase) |
| chr8_+_3543131 | 0.88 |
ENSMUST00000061508.8
ENSMUST00000207318.2 |
Zfp358
|
zinc finger protein 358 |
| chr2_-_77533596 | 0.88 |
ENSMUST00000171063.8
|
Zfp385b
|
zinc finger protein 385B |
| chr16_+_78098626 | 0.88 |
ENSMUST00000114229.4
ENSMUST00000231356.2 |
Cxadr
|
coxsackie virus and adenovirus receptor |
| chr9_-_72018933 | 0.88 |
ENSMUST00000185117.8
|
Tcf12
|
transcription factor 12 |
| chr13_+_83720457 | 0.87 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr3_+_95801325 | 0.87 |
ENSMUST00000197081.2
ENSMUST00000056710.10 |
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chr9_+_117869577 | 0.87 |
ENSMUST00000133580.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr11_-_68277631 | 0.86 |
ENSMUST00000021284.4
|
Ntn1
|
netrin 1 |
| chr3_+_129326004 | 0.86 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr4_-_126427234 | 0.86 |
ENSMUST00000084289.5
|
Ago4
|
argonaute RISC catalytic subunit 4 |
| chr8_-_71315902 | 0.85 |
ENSMUST00000212611.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr18_+_21135079 | 0.84 |
ENSMUST00000234545.2
|
Rnf138
|
ring finger protein 138 |
| chr1_-_14380418 | 0.84 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr17_+_47747657 | 0.84 |
ENSMUST00000150819.3
|
AI661453
|
expressed sequence AI661453 |
| chr9_-_50639367 | 0.83 |
ENSMUST00000117646.8
|
Dixdc1
|
DIX domain containing 1 |
| chr3_+_135531409 | 0.83 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr13_-_40882417 | 0.83 |
ENSMUST00000225180.2
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr6_-_120341304 | 0.82 |
ENSMUST00000146667.2
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr4_+_105014536 | 0.82 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr1_-_14380032 | 0.80 |
ENSMUST00000187790.2
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr9_+_51958453 | 0.80 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr3_+_129326285 | 0.79 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr18_+_35695736 | 0.79 |
ENSMUST00000235851.2
ENSMUST00000235581.2 |
Matr3
|
matrin 3 |
| chr1_-_14380327 | 0.79 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox5_Sry
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 36.0 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 1.8 | 7.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.1 | 3.4 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 1.1 | 9.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 1.1 | 3.2 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.9 | 4.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.9 | 6.3 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.8 | 5.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.7 | 3.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.7 | 5.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 2.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.6 | 1.8 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.6 | 2.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.5 | 1.6 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.5 | 2.6 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.5 | 2.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 1.5 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.5 | 1.5 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.5 | 2.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.5 | 4.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.5 | 3.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.5 | 1.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.5 | 1.4 | GO:0090320 | regulation of chylomicron remnant clearance(GO:0090320) |
| 0.4 | 2.9 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.4 | 1.5 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.4 | 1.8 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.4 | 3.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.4 | 1.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.3 | 2.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 2.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.3 | 3.8 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.3 | 1.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.3 | 3.4 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.3 | 0.9 | GO:0014728 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.3 | 1.5 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.3 | 0.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 0.9 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 1.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 5.5 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 2.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.3 | 1.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.3 | 3.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 0.8 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.3 | 1.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.3 | 2.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 1.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 1.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 3.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 5.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 3.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.9 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 2.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 0.7 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.2 | 2.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 0.7 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 2.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 2.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 2.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 5.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 1.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 2.3 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 0.4 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 0.2 | 0.6 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.2 | 0.6 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.2 | 0.8 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 0.6 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.2 | 1.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 0.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 1.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.9 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.2 | 1.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.2 | 0.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.2 | 0.8 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.2 | 1.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.2 | 2.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.8 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 3.4 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.7 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 1.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 1.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 1.7 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.5 | GO:0098707 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 4.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 1.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 3.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.7 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.6 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.1 | 0.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 5.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 4.3 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.1 | 2.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 0.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.7 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.8 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.3 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 2.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 1.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.3 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.9 | GO:0070922 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) small RNA loading onto RISC(GO:0070922) |
| 0.1 | 9.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.6 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 1.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.4 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.2 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 1.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.8 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 2.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.7 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 1.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.2 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.6 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 1.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 1.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.6 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.4 | GO:0046959 | habituation(GO:0046959) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 1.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.1 | 3.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.2 | GO:0051866 | epithalamus development(GO:0021538) habenula development(GO:0021986) general adaptation syndrome(GO:0051866) |
| 0.1 | 0.2 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.4 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 1.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.7 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 2.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 1.5 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 1.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.3 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 1.0 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 1.5 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.6 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.9 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.1 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 1.2 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0051608 | quaternary ammonium group transport(GO:0015697) histamine transport(GO:0051608) |
| 0.0 | 0.3 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.7 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 1.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 36.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.7 | 10.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.6 | 3.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.4 | 4.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 2.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 7.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 2.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 3.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 2.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.6 | GO:0005712 | chiasma(GO:0005712) |
| 0.2 | 0.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 0.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 2.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 2.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 3.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 3.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.9 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 4.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 8.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 3.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.6 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.9 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.2 | GO:0044317 | rod spherule(GO:0044317) |
| 0.1 | 1.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.7 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.9 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 7.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 3.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 4.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 3.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 1.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 7.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 10.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 4.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.1 | 3.4 | GO:0001761 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 1.0 | 7.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 5.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.8 | 36.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.5 | 8.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.5 | 3.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.5 | 3.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.4 | 1.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 5.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 2.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 2.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 1.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 1.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 0.9 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.3 | 2.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.3 | 1.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 1.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 1.4 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.3 | 1.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 1.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 6.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 2.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 2.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 1.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.8 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 0.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 2.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 0.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 1.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 6.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 4.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 2.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 3.5 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.1 | 0.8 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.3 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 3.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.4 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 3.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 6.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.4 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 1.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.7 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 4.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 4.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.2 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 1.0 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 8.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 5.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 4.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.4 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 3.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 2.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 2.5 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 1.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 3.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 44.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 8.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 10.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 8.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 5.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 5.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 5.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.3 | 7.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 0.7 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.2 | 3.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 5.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.3 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 2.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 6.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 3.7 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 5.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 6.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |