Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
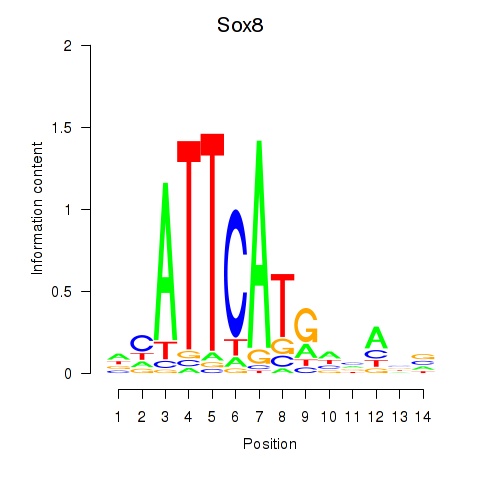
Results for Sox8
Z-value: 0.40
Transcription factors associated with Sox8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox8
|
ENSMUSG00000024176.11 | SRY (sex determining region Y)-box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox8 | mm39_v1_chr17_-_25789652_25789670 | 0.13 | 4.5e-01 | Click! |
Activity profile of Sox8 motif
Sorted Z-values of Sox8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_65008735 | 2.40 |
ENSMUST00000213533.2
ENSMUST00000035499.5 ENSMUST00000077696.13 ENSMUST00000166273.2 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr7_-_126399574 | 1.71 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_100164007 | 1.68 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr1_-_134006847 | 1.66 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr7_-_126399208 | 1.66 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_126399778 | 1.56 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr10_+_11157326 | 1.34 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr5_-_44259010 | 1.26 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr5_-_44259293 | 1.24 |
ENSMUST00000074113.13
|
Prom1
|
prominin 1 |
| chr15_-_103123711 | 0.97 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr2_+_4405769 | 0.88 |
ENSMUST00000075767.14
|
Frmd4a
|
FERM domain containing 4A |
| chr6_-_72357398 | 0.87 |
ENSMUST00000101285.10
ENSMUST00000074231.6 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr4_-_116263183 | 0.85 |
ENSMUST00000123072.8
ENSMUST00000144281.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr8_+_95472218 | 0.84 |
ENSMUST00000034231.4
|
Ccl22
|
chemokine (C-C motif) ligand 22 |
| chr1_+_136059101 | 0.78 |
ENSMUST00000075164.11
|
Kif21b
|
kinesin family member 21B |
| chr11_+_114618209 | 0.75 |
ENSMUST00000069325.14
|
Dnai2
|
dynein axonemal intermediate chain 2 |
| chr11_-_69649452 | 0.68 |
ENSMUST00000058470.16
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr13_-_31134281 | 0.62 |
ENSMUST00000102946.8
|
Exoc2
|
exocyst complex component 2 |
| chrX_+_157993303 | 0.54 |
ENSMUST00000112493.8
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr4_+_11758147 | 0.51 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr2_+_85809620 | 0.47 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr10_+_58207229 | 0.47 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr1_-_79838897 | 0.46 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr11_+_113550109 | 0.39 |
ENSMUST00000137878.2
|
Cog1
|
component of oligomeric golgi complex 1 |
| chrX_-_23151771 | 0.39 |
ENSMUST00000115319.9
|
Klhl13
|
kelch-like 13 |
| chr12_-_115722081 | 0.34 |
ENSMUST00000103541.3
|
Ighv1-72
|
immunoglobulin heavy variable 1-72 |
| chr10_-_128016135 | 0.33 |
ENSMUST00000238843.2
ENSMUST00000099139.9 |
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr14_-_8457069 | 0.30 |
ENSMUST00000022257.4
|
Atxn7
|
ataxin 7 |
| chr19_-_33764859 | 0.28 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr19_-_40365318 | 0.27 |
ENSMUST00000239304.2
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr12_-_114955196 | 0.25 |
ENSMUST00000194865.2
|
Ighv1-47
|
immunoglobulin heavy variable 1-47 |
| chr9_+_109881083 | 0.24 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chr6_-_58452315 | 0.24 |
ENSMUST00000177318.3
ENSMUST00000176147.9 ENSMUST00000176023.3 ENSMUST00000228586.2 |
Vmn1r31
|
vomeronasal 1 receptor 31 |
| chr2_+_109522781 | 0.23 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr9_-_95697441 | 0.22 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr12_-_115083839 | 0.21 |
ENSMUST00000103521.3
|
Ighv1-50
|
immunoglobulin heavy variable 1-50 |
| chr12_-_115425105 | 0.20 |
ENSMUST00000103532.3
|
Ighv1-62-3
|
immunoglobulin heavy variable 1-62-3 |
| chr13_+_19528728 | 0.20 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr13_+_93441307 | 0.17 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr1_+_135710803 | 0.17 |
ENSMUST00000132795.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr14_+_21881794 | 0.15 |
ENSMUST00000152562.8
|
Vdac2
|
voltage-dependent anion channel 2 |
| chr18_+_37637317 | 0.15 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr17_-_26052335 | 0.15 |
ENSMUST00000044911.10
ENSMUST00000237186.2 |
Stub1
|
STIP1 homology and U-Box containing protein 1 |
| chrX_-_71961890 | 0.14 |
ENSMUST00000152200.2
|
Cetn2
|
centrin 2 |
| chrX_+_133305529 | 0.13 |
ENSMUST00000113224.9
ENSMUST00000113226.2 |
Drp2
|
dystrophin related protein 2 |
| chr11_-_99884818 | 0.13 |
ENSMUST00000105049.2
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr7_+_102423702 | 0.13 |
ENSMUST00000098217.3
|
Olfr561
|
olfactory receptor 561 |
| chr1_+_109911467 | 0.09 |
ENSMUST00000172005.8
|
Cdh7
|
cadherin 7, type 2 |
| chr18_+_65831324 | 0.09 |
ENSMUST00000115097.8
ENSMUST00000117694.2 ENSMUST00000235962.2 |
Oacyl
|
O-acyltransferase like |
| chr16_-_29360301 | 0.07 |
ENSMUST00000057018.15
ENSMUST00000182627.8 |
Atp13a4
|
ATPase type 13A4 |
| chr4_+_154226819 | 0.07 |
ENSMUST00000030895.12
|
Wrap73
|
WD repeat containing, antisense to Trp73 |
| chr11_+_69882410 | 0.06 |
ENSMUST00000108592.2
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr6_-_42524521 | 0.06 |
ENSMUST00000217978.2
|
Olfr455
|
olfactory receptor 455 |
| chr12_-_114843941 | 0.05 |
ENSMUST00000191862.6
ENSMUST00000103513.3 |
Ighv1-36
|
immunoglobulin heavy variable 1-36 |
| chrX_-_49517375 | 0.05 |
ENSMUST00000215270.2
|
Olfr1324
|
olfactory receptor 1324 |
| chr18_+_37085673 | 0.05 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr3_-_92393193 | 0.05 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr4_+_127062924 | 0.04 |
ENSMUST00000046659.14
|
Dlgap3
|
DLG associated protein 3 |
| chr11_-_43638790 | 0.04 |
ENSMUST00000048578.3
ENSMUST00000109278.8 |
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr4_-_133972890 | 0.04 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr12_-_115172211 | 0.04 |
ENSMUST00000103526.3
|
Ighv1-55
|
immunoglobulin heavy variable 1-55 |
| chr12_+_113038376 | 0.03 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chr12_-_114451189 | 0.03 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr13_-_19579898 | 0.03 |
ENSMUST00000197565.3
ENSMUST00000221380.2 ENSMUST00000200323.3 ENSMUST00000199924.2 ENSMUST00000222869.2 |
Stard3nl
|
STARD3 N-terminal like |
| chr6_+_41039255 | 0.03 |
ENSMUST00000103266.3
|
Trbv5
|
T cell receptor beta, variable 5 |
| chr11_-_46057224 | 0.02 |
ENSMUST00000020679.3
|
Nipal4
|
NIPA-like domain containing 4 |
| chr5_+_43672856 | 0.02 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr4_-_129472328 | 0.01 |
ENSMUST00000052835.9
|
Fam167b
|
family with sequence similarity 167, member B |
| chr3_+_101917392 | 0.00 |
ENSMUST00000196324.2
|
Nhlh2
|
nescient helix loop helix 2 |
| chr7_+_119773070 | 0.00 |
ENSMUST00000033201.7
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.4 | 1.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 4.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 0.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 0.5 | GO:0042628 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) seminal vesicle development(GO:0061107) |
| 0.1 | 0.6 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 1.7 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.9 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.7 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 4.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 1.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.6 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.5 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 2.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 1.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |