Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
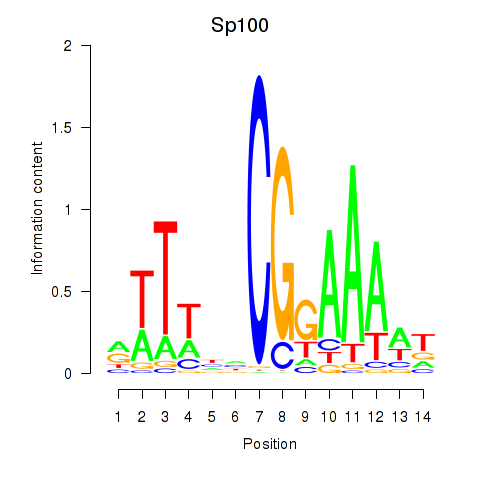
Results for Sp100
Z-value: 0.94
Transcription factors associated with Sp100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp100
|
ENSMUSG00000026222.17 | nuclear antigen Sp100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | mm39_v1_chr1_+_85577709_85577740 | 0.06 | 7.2e-01 | Click! |
Activity profile of Sp100 motif
Sorted Z-values of Sp100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_94905710 | 11.91 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr14_+_66043515 | 4.41 |
ENSMUST00000139644.2
|
Pbk
|
PDZ binding kinase |
| chr16_-_16942970 | 3.98 |
ENSMUST00000093336.8
ENSMUST00000231681.2 |
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr9_-_21874802 | 3.88 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr4_-_132260799 | 3.63 |
ENSMUST00000152993.8
ENSMUST00000067496.7 |
Atpif1
|
ATPase inhibitory factor 1 |
| chr8_-_4829519 | 3.57 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr6_+_18848570 | 3.56 |
ENSMUST00000056398.11
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr17_-_7050145 | 3.51 |
ENSMUST00000064234.7
|
Ezr
|
ezrin |
| chr3_+_134918298 | 3.32 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr17_+_47816074 | 3.20 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr8_+_118225008 | 3.01 |
ENSMUST00000081232.9
|
Plcg2
|
phospholipase C, gamma 2 |
| chr15_-_98832403 | 2.89 |
ENSMUST00000077577.8
|
Tuba1b
|
tubulin, alpha 1B |
| chr16_+_48692976 | 2.89 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr13_+_117356808 | 2.84 |
ENSMUST00000022242.9
|
Emb
|
embigin |
| chr7_+_13467422 | 2.83 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr5_-_21990170 | 2.72 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chrX_-_142610371 | 2.57 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr16_-_21980200 | 2.45 |
ENSMUST00000115379.2
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr18_-_62313019 | 2.38 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr5_-_44259010 | 2.33 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr4_+_108436639 | 2.32 |
ENSMUST00000102744.4
|
Orc1
|
origin recognition complex, subunit 1 |
| chr13_-_49806231 | 2.25 |
ENSMUST00000021818.9
|
Cenpp
|
centromere protein P |
| chr3_+_122068018 | 2.23 |
ENSMUST00000035776.10
|
Dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr17_+_47816137 | 2.17 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr2_-_98497609 | 2.10 |
ENSMUST00000099683.2
|
Gm10800
|
predicted gene 10800 |
| chrX_-_165992145 | 2.07 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr10_-_88520877 | 1.97 |
ENSMUST00000138734.2
|
Spic
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr2_+_75489596 | 1.95 |
ENSMUST00000111964.8
ENSMUST00000111962.8 ENSMUST00000111961.8 ENSMUST00000164947.9 ENSMUST00000090792.11 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr6_-_131270136 | 1.88 |
ENSMUST00000032307.12
|
Magohb
|
mago homolog B, exon junction complex core component |
| chr3_-_157742267 | 1.83 |
ENSMUST00000121326.8
|
Srsf11
|
serine and arginine-rich splicing factor 11 |
| chr6_+_18848600 | 1.80 |
ENSMUST00000201141.3
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr2_+_91376650 | 1.76 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr3_-_157742182 | 1.74 |
ENSMUST00000069025.14
|
Srsf11
|
serine and arginine-rich splicing factor 11 |
| chr16_-_4536943 | 1.70 |
ENSMUST00000120056.8
ENSMUST00000074970.8 |
Nmral1
|
NmrA-like family domain containing 1 |
| chr6_-_87649173 | 1.68 |
ENSMUST00000032130.8
|
Aplf
|
aprataxin and PNKP like factor |
| chr4_+_33310306 | 1.65 |
ENSMUST00000108153.9
ENSMUST00000029942.8 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr17_-_36149100 | 1.62 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr15_+_85743887 | 1.62 |
ENSMUST00000170629.3
|
Gtse1
|
G two S phase expressed protein 1 |
| chr11_+_22940599 | 1.61 |
ENSMUST00000020562.5
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr18_+_11790409 | 1.59 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr7_-_99770653 | 1.55 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr8_-_65489834 | 1.55 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr2_-_168609110 | 1.52 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr12_-_28685849 | 1.50 |
ENSMUST00000221871.2
|
Rps7
|
ribosomal protein S7 |
| chr11_-_49603501 | 1.50 |
ENSMUST00000020624.7
ENSMUST00000145353.8 |
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr1_-_178165223 | 1.48 |
ENSMUST00000037748.9
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U |
| chr6_-_87649124 | 1.47 |
ENSMUST00000065997.5
|
Aplf
|
aprataxin and PNKP like factor |
| chr9_+_120400510 | 1.42 |
ENSMUST00000165532.3
|
Rpl14
|
ribosomal protein L14 |
| chr3_+_103767581 | 1.38 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr9_+_26941453 | 1.36 |
ENSMUST00000073127.14
ENSMUST00000086198.5 |
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr17_-_10501816 | 1.34 |
ENSMUST00000233684.2
|
Qk
|
quaking, KH domain containing RNA binding |
| chr9_+_3023547 | 1.31 |
ENSMUST00000099046.4
|
Gm10718
|
predicted gene 10718 |
| chr8_+_70735477 | 1.31 |
ENSMUST00000087467.12
ENSMUST00000140212.8 ENSMUST00000110124.9 |
Homer3
|
homer scaffolding protein 3 |
| chr13_-_23946359 | 1.30 |
ENSMUST00000091701.3
|
H3c1
|
H3 clustered histone 1 |
| chr10_+_25284244 | 1.29 |
ENSMUST00000092645.7
ENSMUST00000219805.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr12_-_28685913 | 1.28 |
ENSMUST00000074267.5
|
Rps7
|
ribosomal protein S7 |
| chr3_-_157742204 | 1.27 |
ENSMUST00000137444.8
|
Srsf11
|
serine and arginine-rich splicing factor 11 |
| chrX_+_55825033 | 1.26 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr9_+_26941945 | 1.26 |
ENSMUST00000216677.2
|
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr14_-_57015748 | 1.25 |
ENSMUST00000022507.13
ENSMUST00000163924.2 |
Pspc1
|
paraspeckle protein 1 |
| chr4_-_126096551 | 1.25 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr9_+_56325893 | 1.25 |
ENSMUST00000034879.5
ENSMUST00000215269.2 |
Hmg20a
|
high mobility group 20A |
| chrX_+_132959905 | 1.21 |
ENSMUST00000113287.8
ENSMUST00000033609.9 ENSMUST00000113286.8 |
Cstf2
|
cleavage stimulation factor, 3' pre-RNA subunit 2 |
| chrX_-_165992311 | 1.17 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr7_-_13856967 | 1.17 |
ENSMUST00000098809.4
|
Sult2a3
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 3 |
| chr13_+_38223023 | 1.16 |
ENSMUST00000226110.2
|
Riok1
|
RIO kinase 1 |
| chr4_-_126096376 | 1.15 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr5_+_129864044 | 1.15 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr15_-_96597610 | 1.11 |
ENSMUST00000023099.8
|
Slc38a2
|
solute carrier family 38, member 2 |
| chr1_+_82702598 | 1.10 |
ENSMUST00000078332.13
ENSMUST00000073025.12 ENSMUST00000161648.8 ENSMUST00000160786.8 ENSMUST00000162003.8 |
Mff
|
mitochondrial fission factor |
| chr12_-_75678092 | 1.09 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr17_+_47922497 | 1.08 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr9_-_36637670 | 1.07 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr7_+_13357892 | 1.06 |
ENSMUST00000108525.4
|
Sult2a5
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 5 |
| chrX_+_55824797 | 1.05 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr8_-_65489791 | 1.05 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr9_-_21671571 | 1.03 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr15_+_12205135 | 1.03 |
ENSMUST00000071993.13
|
Mtmr12
|
myotubularin related protein 12 |
| chr14_+_4198620 | 1.03 |
ENSMUST00000080281.14
|
Rpl15
|
ribosomal protein L15 |
| chr11_+_72332167 | 1.02 |
ENSMUST00000045633.6
|
Mybbp1a
|
MYB binding protein (P160) 1a |
| chr15_-_81810349 | 1.02 |
ENSMUST00000023113.7
|
Polr3h
|
polymerase (RNA) III (DNA directed) polypeptide H |
| chr1_-_118239146 | 1.00 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr8_-_31658775 | 0.99 |
ENSMUST00000033983.6
|
Mak16
|
MAK16 homolog |
| chr5_-_129864202 | 0.99 |
ENSMUST00000136507.4
|
Psph
|
phosphoserine phosphatase |
| chr1_-_60137294 | 0.95 |
ENSMUST00000141417.3
ENSMUST00000122038.8 |
Wdr12
|
WD repeat domain 12 |
| chr12_-_15866763 | 0.94 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr18_-_64649497 | 0.93 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr15_-_64254754 | 0.93 |
ENSMUST00000177374.8
ENSMUST00000110114.10 ENSMUST00000110115.9 ENSMUST00000023008.16 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr12_-_69403892 | 0.93 |
ENSMUST00000222065.2
ENSMUST00000021368.10 |
Nemf
|
nuclear export mediator factor |
| chr7_+_27869115 | 0.92 |
ENSMUST00000042405.8
|
Fbl
|
fibrillarin |
| chr18_-_64649620 | 0.92 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr11_-_96668318 | 0.91 |
ENSMUST00000127375.2
ENSMUST00000021246.9 ENSMUST00000107661.10 |
Snx11
|
sorting nexin 11 |
| chr2_+_70886073 | 0.91 |
ENSMUST00000154704.8
ENSMUST00000135357.8 ENSMUST00000064141.12 ENSMUST00000112159.9 ENSMUST00000102701.5 |
Dcaf17
|
DDB1 and CUL4 associated factor 17 |
| chr4_-_58987094 | 0.91 |
ENSMUST00000030069.7
|
Ptgr1
|
prostaglandin reductase 1 |
| chr16_-_19801781 | 0.90 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chrX_+_36390430 | 0.89 |
ENSMUST00000016553.5
|
Nkap
|
NFKB activating protein |
| chr16_+_20536415 | 0.88 |
ENSMUST00000021405.8
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr7_-_3723381 | 0.88 |
ENSMUST00000078451.7
|
Pirb
|
paired Ig-like receptor B |
| chr7_+_128346655 | 0.86 |
ENSMUST00000042942.10
|
Sec23ip
|
Sec23 interacting protein |
| chr9_+_3335469 | 0.86 |
ENSMUST00000053407.13
ENSMUST00000211933.2 ENSMUST00000212666.2 |
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr9_-_36637923 | 0.84 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr11_-_78642480 | 0.83 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr18_+_12261798 | 0.83 |
ENSMUST00000025270.8
|
Riok3
|
RIO kinase 3 |
| chr15_+_12205095 | 0.82 |
ENSMUST00000038172.16
|
Mtmr12
|
myotubularin related protein 12 |
| chr11_-_115915315 | 0.81 |
ENSMUST00000016703.8
|
H3f3b
|
H3.3 histone B |
| chr4_-_135749032 | 0.80 |
ENSMUST00000030427.6
|
Eloa
|
elongin A |
| chr11_-_16458069 | 0.78 |
ENSMUST00000109641.2
|
Sec61g
|
SEC61, gamma subunit |
| chr11_-_59855762 | 0.77 |
ENSMUST00000062405.8
|
Rasd1
|
RAS, dexamethasone-induced 1 |
| chr9_-_30833748 | 0.75 |
ENSMUST00000065112.7
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr2_+_84867554 | 0.74 |
ENSMUST00000077798.13
|
Ssrp1
|
structure specific recognition protein 1 |
| chr2_-_105833894 | 0.74 |
ENSMUST00000102555.11
|
Dnajc24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr2_+_71042050 | 0.74 |
ENSMUST00000112142.8
ENSMUST00000112139.8 ENSMUST00000112140.8 ENSMUST00000112138.8 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr8_-_48128164 | 0.73 |
ENSMUST00000080353.3
|
Ing2
|
inhibitor of growth family, member 2 |
| chr6_+_135339929 | 0.73 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr13_-_46881388 | 0.72 |
ENSMUST00000021803.10
|
Nup153
|
nucleoporin 153 |
| chrX_+_20570145 | 0.71 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr11_-_16458484 | 0.70 |
ENSMUST00000109643.8
ENSMUST00000166950.8 ENSMUST00000178855.8 |
Sec61g
|
SEC61, gamma subunit |
| chr15_-_64254585 | 0.70 |
ENSMUST00000176384.8
ENSMUST00000175799.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr9_+_3000922 | 0.68 |
ENSMUST00000151376.3
|
Gm10722
|
predicted gene 10722 |
| chr11_-_110058899 | 0.67 |
ENSMUST00000044850.4
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr6_+_21985902 | 0.67 |
ENSMUST00000115383.9
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_+_4795873 | 0.66 |
ENSMUST00000032597.12
ENSMUST00000078432.5 |
Rpl28
|
ribosomal protein L28 |
| chr13_+_104953679 | 0.66 |
ENSMUST00000022230.15
|
Srek1ip1
|
splicing regulatory glutamine/lysine-rich protein 1interacting protein 1 |
| chr9_+_3013140 | 0.65 |
ENSMUST00000143083.3
|
Gm10721
|
predicted gene 10721 |
| chr8_-_106198112 | 0.64 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr2_+_109848224 | 0.63 |
ENSMUST00000150183.9
|
Ccdc34
|
coiled-coil domain containing 34 |
| chr5_+_21990251 | 0.63 |
ENSMUST00000239497.2
ENSMUST00000030769.7 |
Psmc2
|
proteasome (prosome, macropain) 26S subunit, ATPase 2 |
| chr17_-_47922374 | 0.63 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr4_+_129714494 | 0.62 |
ENSMUST00000165853.2
|
Ptp4a2
|
protein tyrosine phosphatase 4a2 |
| chr8_+_31640332 | 0.62 |
ENSMUST00000209851.2
ENSMUST00000098842.3 ENSMUST00000210129.2 |
Tti2
|
TELO2 interacting protein 2 |
| chr13_+_23930717 | 0.61 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr2_-_151583155 | 0.61 |
ENSMUST00000042452.11
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr7_+_101546059 | 0.60 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr8_-_25592385 | 0.60 |
ENSMUST00000064883.14
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr18_-_67582191 | 0.60 |
ENSMUST00000025408.10
|
Afg3l2
|
AFG3-like AAA ATPase 2 |
| chr14_-_51162346 | 0.59 |
ENSMUST00000159292.8
|
Osgep
|
O-sialoglycoprotein endopeptidase |
| chr18_+_32970278 | 0.59 |
ENSMUST00000053663.11
|
Wdr36
|
WD repeat domain 36 |
| chr12_+_31704853 | 0.59 |
ENSMUST00000036862.5
|
Cog5
|
component of oligomeric golgi complex 5 |
| chr4_-_3835595 | 0.59 |
ENSMUST00000138502.2
|
Rps20
|
ribosomal protein S20 |
| chr11_+_97206542 | 0.58 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr4_-_108436514 | 0.58 |
ENSMUST00000079213.6
|
Prpf38a
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing A |
| chr16_+_20536545 | 0.58 |
ENSMUST00000231656.2
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr5_+_146885450 | 0.57 |
ENSMUST00000146511.8
ENSMUST00000132102.2 |
Gtf3a
|
general transcription factor III A |
| chr7_-_12096691 | 0.57 |
ENSMUST00000086228.3
|
Vmn1r84
|
vomeronasal 1 receptor 84 |
| chr7_-_143102524 | 0.57 |
ENSMUST00000208093.2
ENSMUST00000209098.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr5_+_150597204 | 0.55 |
ENSMUST00000202170.4
ENSMUST00000016569.11 |
Pds5b
|
PDS5 cohesin associated factor B |
| chr13_-_108295018 | 0.54 |
ENSMUST00000225830.2
ENSMUST00000223734.2 ENSMUST00000163558.3 |
Ndufaf2
|
NADH:ubiquinone oxidoreductase complex assembly factor 2 |
| chr8_-_57003828 | 0.54 |
ENSMUST00000134162.8
ENSMUST00000140107.8 ENSMUST00000040330.15 ENSMUST00000135337.8 |
Cep44
|
centrosomal protein 44 |
| chr1_+_40363701 | 0.53 |
ENSMUST00000095020.9
ENSMUST00000194296.6 |
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr15_+_101982208 | 0.53 |
ENSMUST00000169681.3
ENSMUST00000229400.2 |
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr14_-_70405288 | 0.52 |
ENSMUST00000129174.8
ENSMUST00000125300.3 |
Pdlim2
|
PDZ and LIM domain 2 |
| chr2_+_127050281 | 0.52 |
ENSMUST00000103220.4
|
Snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr13_+_17869988 | 0.51 |
ENSMUST00000049744.4
|
Mplkip
|
M-phase specific PLK1 intereacting protein |
| chr5_+_138228074 | 0.51 |
ENSMUST00000159798.8
ENSMUST00000159964.2 |
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr4_-_129590609 | 0.51 |
ENSMUST00000102588.10
|
Tmem39b
|
transmembrane protein 39b |
| chr10_-_112764879 | 0.50 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr4_-_129517662 | 0.49 |
ENSMUST00000106035.8
ENSMUST00000150357.2 ENSMUST00000030586.15 |
Ccdc28b
|
coiled coil domain containing 28B |
| chr8_+_71126012 | 0.49 |
ENSMUST00000146972.3
ENSMUST00000210071.2 ENSMUST00000210987.2 |
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_-_55612172 | 0.46 |
ENSMUST00000126604.8
ENSMUST00000117812.8 ENSMUST00000119201.8 |
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 homolog (human) |
| chr11_-_70242850 | 0.46 |
ENSMUST00000019068.7
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr4_-_124587340 | 0.45 |
ENSMUST00000030738.8
|
Utp11
|
UTP11 small subunit processome component |
| chr5_+_136164990 | 0.45 |
ENSMUST00000136634.8
ENSMUST00000041100.4 |
Alkbh4
|
alkB homolog 4, lysine demethylase |
| chr5_-_21156766 | 0.45 |
ENSMUST00000036489.10
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr2_+_71042172 | 0.44 |
ENSMUST00000081710.12
|
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr2_+_71042285 | 0.44 |
ENSMUST00000112144.9
ENSMUST00000100028.10 ENSMUST00000112136.2 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr7_-_3551003 | 0.43 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr2_+_122478882 | 0.42 |
ENSMUST00000142767.8
|
AA467197
|
expressed sequence AA467197 |
| chr2_+_119727689 | 0.42 |
ENSMUST00000046717.13
ENSMUST00000079934.12 ENSMUST00000110774.8 ENSMUST00000110773.9 ENSMUST00000156510.2 |
Mga
|
MAX gene associated |
| chr18_+_32970363 | 0.42 |
ENSMUST00000166214.9
|
Wdr36
|
WD repeat domain 36 |
| chr3_-_59038160 | 0.42 |
ENSMUST00000197841.2
|
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr1_-_173703424 | 0.42 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr11_-_16458148 | 0.41 |
ENSMUST00000109642.8
|
Sec61g
|
SEC61, gamma subunit |
| chr19_-_56996617 | 0.41 |
ENSMUST00000118800.8
ENSMUST00000111584.9 ENSMUST00000122359.8 ENSMUST00000148049.8 |
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr7_-_13723513 | 0.41 |
ENSMUST00000165167.8
ENSMUST00000108520.4 |
Sult2a4
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 4 |
| chr4_+_155709275 | 0.41 |
ENSMUST00000067081.10
ENSMUST00000105600.8 ENSMUST00000105598.2 |
Cdk11b
|
cyclin-dependent kinase 11B |
| chr9_-_37525009 | 0.41 |
ENSMUST00000002013.11
|
Spa17
|
sperm autoantigenic protein 17 |
| chr12_-_31704766 | 0.40 |
ENSMUST00000020977.4
|
Dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr16_-_14135183 | 0.40 |
ENSMUST00000120707.2
ENSMUST00000023357.14 |
Cep20
|
centrosomal protein 20 |
| chr13_+_21364330 | 0.39 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr5_+_31771281 | 0.39 |
ENSMUST00000031024.14
ENSMUST00000202033.2 |
Mrpl33
Gm43809
|
mitochondrial ribosomal protein L33 predicted gene 43809 |
| chr13_+_109397184 | 0.39 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr2_-_163913014 | 0.39 |
ENSMUST00000018466.4
ENSMUST00000109384.10 |
Tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr3_-_116456292 | 0.38 |
ENSMUST00000029570.9
|
Mfsd14a
|
major facilitator superfamily domain containing 14A |
| chr9_+_3403592 | 0.38 |
ENSMUST00000027027.7
|
Cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr2_-_88810595 | 0.38 |
ENSMUST00000168169.10
ENSMUST00000144908.3 |
Olfr1213
|
olfactory receptor 1213 |
| chr8_-_71349927 | 0.37 |
ENSMUST00000212709.2
ENSMUST00000212796.2 ENSMUST00000212378.2 ENSMUST00000054220.10 ENSMUST00000212494.2 |
Rpl18a
|
ribosomal protein L18A |
| chr16_+_33201164 | 0.37 |
ENSMUST00000165418.9
|
Zfp148
|
zinc finger protein 148 |
| chr15_-_85695855 | 0.37 |
ENSMUST00000134631.8
ENSMUST00000154814.2 ENSMUST00000071876.13 ENSMUST00000150995.8 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr9_+_3017408 | 0.36 |
ENSMUST00000099049.4
|
Gm10719
|
predicted gene 10719 |
| chr14_+_73475335 | 0.35 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr6_-_28421678 | 0.34 |
ENSMUST00000090511.4
|
Gcc1
|
golgi coiled coil 1 |
| chr2_+_136734088 | 0.34 |
ENSMUST00000099311.9
|
Slx4ip
|
SLX4 interacting protein |
| chr15_+_41573995 | 0.34 |
ENSMUST00000229769.2
|
Oxr1
|
oxidation resistance 1 |
| chr7_+_126376099 | 0.33 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr11_-_78067446 | 0.33 |
ENSMUST00000148154.3
ENSMUST00000017549.13 |
Nek8
|
NIMA (never in mitosis gene a)-related expressed kinase 8 |
| chr5_-_124490296 | 0.33 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr10_+_82696135 | 0.32 |
ENSMUST00000219442.3
|
Txnrd1
|
thioredoxin reductase 1 |
| chr5_+_16758538 | 0.32 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor |
| chr13_-_104365310 | 0.31 |
ENSMUST00000022226.6
|
Ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr6_+_134958681 | 0.31 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp100
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 11.9 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.9 | 3.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.8 | 2.3 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.7 | 2.9 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.6 | 1.9 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.6 | 3.6 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.6 | 2.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.5 | 2.3 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 1.4 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.5 | 3.2 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.4 | 3.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.4 | 2.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 1.5 | GO:2000371 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.4 | 1.9 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.4 | 3.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.3 | 1.0 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.3 | 0.9 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.3 | 1.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 1.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 2.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 3.9 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.3 | 2.8 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.2 | 0.7 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 2.8 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 1.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 5.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 1.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 3.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 1.6 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 1.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.0 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 1.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 0.8 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 0.5 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.2 | 0.5 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.6 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.8 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 1.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.2 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.1 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 1.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 1.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.2 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.1 | 1.1 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.1 | 0.3 | GO:1904172 | positive regulation of bleb assembly(GO:1904172) |
| 0.1 | 1.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 | 0.7 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 1.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 2.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.3 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 1.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 4.3 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 2.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 2.7 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.6 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 1.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 2.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 2.1 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 2.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 1.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0090306 | polar body extrusion after meiotic divisions(GO:0040038) spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.6 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.2 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 4.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.8 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 3.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.9 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.7 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.3 | GO:2000169 | cellular response to hydroperoxide(GO:0071447) negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.8 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 5.4 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 1.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 1.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.3 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.4 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.9 | GO:0051168 | nuclear export(GO:0051168) |
| 0.0 | 1.1 | GO:0019236 | response to pheromone(GO:0019236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.5 | 2.6 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.4 | 5.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.3 | 1.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.3 | 2.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 1.2 | GO:0071920 | cleavage body(GO:0071920) |
| 0.2 | 1.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 3.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 2.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 2.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.6 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 3.6 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 4.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.8 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 2.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 3.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 5.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.6 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 5.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 2.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 3.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 6.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.6 | 2.4 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.6 | 1.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 2.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.5 | 1.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.5 | 3.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 1.9 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.4 | 2.6 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.3 | 1.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.3 | 2.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.3 | 2.8 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 1.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 0.9 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 0.6 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.2 | 1.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 1.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 3.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 11.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 2.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 3.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 1.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 5.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.3 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.3 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 3.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.2 | GO:0050145 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 4.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.9 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 2.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 2.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 2.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 4.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 3.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 5.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 3.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 4.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 2.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 1.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 6.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 5.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 2.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 3.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.1 | 2.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 8.8 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 3.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 3.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 4.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 3.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.2 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |