Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
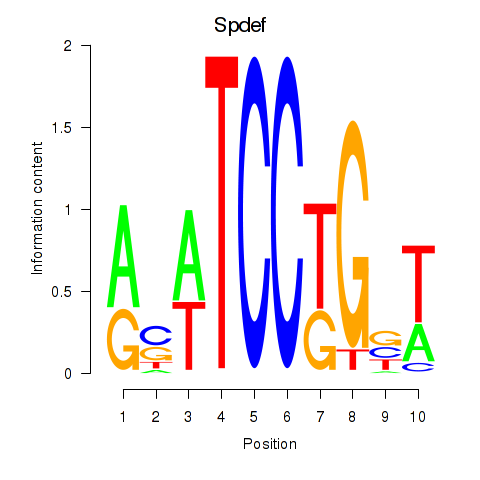
Results for Spdef
Z-value: 0.54
Transcription factors associated with Spdef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spdef
|
ENSMUSG00000024215.15 | SAM pointed domain containing ets transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spdef | mm39_v1_chr17_-_27947863_27947930 | 0.01 | 9.4e-01 | Click! |
Activity profile of Spdef motif
Sorted Z-values of Spdef motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_65815958 | 1.22 |
ENSMUST00000119245.8
ENSMUST00000134338.8 ENSMUST00000179395.8 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr7_+_86895851 | 0.80 |
ENSMUST00000032781.14
|
Nox4
|
NADPH oxidase 4 |
| chrX_-_74886623 | 0.79 |
ENSMUST00000114057.8
|
Pls3
|
plastin 3 (T-isoform) |
| chrX_-_74886791 | 0.78 |
ENSMUST00000137192.2
|
Pls3
|
plastin 3 (T-isoform) |
| chr7_+_45354512 | 0.74 |
ENSMUST00000080885.12
ENSMUST00000211513.2 ENSMUST00000211357.2 |
Dbp
|
D site albumin promoter binding protein |
| chr18_+_67338437 | 0.67 |
ENSMUST00000210564.3
|
Chmp1b
|
charged multivesicular body protein 1B |
| chr2_-_73216743 | 0.62 |
ENSMUST00000112044.8
ENSMUST00000112043.8 ENSMUST00000076463.12 |
Gpr155
|
G protein-coupled receptor 155 |
| chr7_+_130633776 | 0.58 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr3_+_135531548 | 0.57 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr8_+_46080746 | 0.56 |
ENSMUST00000145458.9
ENSMUST00000134321.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr3_-_84489783 | 0.55 |
ENSMUST00000107687.9
ENSMUST00000098990.10 |
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr13_-_59970885 | 0.55 |
ENSMUST00000225179.2
ENSMUST00000225576.2 ENSMUST00000071703.6 |
Tut7
|
terminal uridylyl transferase 7 |
| chr9_+_117869543 | 0.55 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr18_+_12776358 | 0.55 |
ENSMUST00000234966.2
ENSMUST00000025294.9 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr3_-_84489923 | 0.54 |
ENSMUST00000143514.3
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr16_-_87229485 | 0.54 |
ENSMUST00000039449.9
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr9_+_117869577 | 0.52 |
ENSMUST00000133580.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr3_-_5641171 | 0.51 |
ENSMUST00000071280.8
ENSMUST00000195855.6 ENSMUST00000165309.8 ENSMUST00000164828.8 |
Pex2
|
peroxisomal biogenesis factor 2 |
| chr1_+_135746330 | 0.51 |
ENSMUST00000038760.10
|
Lad1
|
ladinin |
| chr18_-_46730547 | 0.51 |
ENSMUST00000151189.2
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr6_-_95695781 | 0.51 |
ENSMUST00000204224.3
|
Suclg2
|
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
| chr3_-_108053396 | 0.50 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr18_-_31742946 | 0.48 |
ENSMUST00000060396.7
|
Slc25a46
|
solute carrier family 25, member 46 |
| chr8_+_114362419 | 0.47 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr5_+_124577952 | 0.46 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr7_-_30672747 | 0.46 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr6_+_90443293 | 0.45 |
ENSMUST00000203607.2
|
Klf15
|
Kruppel-like factor 15 |
| chr2_+_83474779 | 0.45 |
ENSMUST00000081591.7
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr1_+_128171859 | 0.44 |
ENSMUST00000027592.6
|
Ubxn4
|
UBX domain protein 4 |
| chr3_+_135531409 | 0.44 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr9_+_117869642 | 0.44 |
ENSMUST00000134433.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr3_-_5641295 | 0.44 |
ENSMUST00000059021.10
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr16_-_87229367 | 0.43 |
ENSMUST00000232095.2
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr9_+_80072274 | 0.43 |
ENSMUST00000035889.15
ENSMUST00000113268.8 |
Myo6
|
myosin VI |
| chr5_+_67464284 | 0.42 |
ENSMUST00000113676.6
ENSMUST00000162372.8 |
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr18_+_34994253 | 0.42 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr11_+_87482971 | 0.42 |
ENSMUST00000103179.10
ENSMUST00000092802.12 ENSMUST00000146871.8 |
Mtmr4
|
myotubularin related protein 4 |
| chr2_+_71884943 | 0.41 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_-_12424212 | 0.41 |
ENSMUST00000124603.8
ENSMUST00000129993.3 ENSMUST00000028105.13 |
Mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr13_+_41040657 | 0.41 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr3_-_63872189 | 0.41 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr9_+_80072361 | 0.40 |
ENSMUST00000184480.8
|
Myo6
|
myosin VI |
| chr13_-_104057016 | 0.39 |
ENSMUST00000022222.12
|
Erbin
|
Erbb2 interacting protein |
| chrX_-_106859842 | 0.39 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr3_-_82811269 | 0.39 |
ENSMUST00000029632.7
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr9_+_123358810 | 0.39 |
ENSMUST00000026270.9
ENSMUST00000239083.2 |
Sacm1l
|
SAC1 suppressor of actin mutations 1-like (yeast) |
| chr10_+_58207229 | 0.39 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_-_38637474 | 0.38 |
ENSMUST00000143758.8
ENSMUST00000156272.8 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr15_+_35371300 | 0.38 |
ENSMUST00000048646.9
|
Vps13b
|
vacuolar protein sorting 13B |
| chr3_-_63872079 | 0.38 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr1_-_183078488 | 0.38 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr6_-_119365632 | 0.38 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr8_+_114362181 | 0.37 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated |
| chr2_+_145627900 | 0.37 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr18_-_46730381 | 0.37 |
ENSMUST00000036030.14
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr11_-_21522193 | 0.37 |
ENSMUST00000102874.11
ENSMUST00000238916.2 |
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr1_-_84817000 | 0.37 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr11_-_5492175 | 0.37 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr9_-_106353792 | 0.36 |
ENSMUST00000214682.2
ENSMUST00000112479.9 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr8_-_84059048 | 0.36 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr1_+_138891447 | 0.36 |
ENSMUST00000168527.8
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr2_+_31462659 | 0.36 |
ENSMUST00000113482.8
|
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr4_-_4793274 | 0.35 |
ENSMUST00000084949.3
|
Bpnt2
|
3'(2'), 5'-bisphosphate nucleotidase 2 |
| chr5_-_146157711 | 0.35 |
ENSMUST00000169407.9
ENSMUST00000161331.8 ENSMUST00000159074.3 ENSMUST00000067837.10 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr11_-_21521934 | 0.35 |
ENSMUST00000239073.2
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr9_+_123358913 | 0.35 |
ENSMUST00000238984.2
|
Sacm1l
|
SAC1 suppressor of actin mutations 1-like (yeast) |
| chr9_+_110306020 | 0.35 |
ENSMUST00000198858.5
|
Kif9
|
kinesin family member 9 |
| chrX_+_37278636 | 0.34 |
ENSMUST00000049740.3
ENSMUST00000115142.3 ENSMUST00000131124.2 |
Zbtb33
|
zinc finger and BTB domain containing 33 |
| chr17_-_57554631 | 0.34 |
ENSMUST00000233568.2
ENSMUST00000005975.8 |
Gpr108
|
G protein-coupled receptor 108 |
| chr5_+_138083345 | 0.33 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr9_+_110306052 | 0.33 |
ENSMUST00000197248.5
ENSMUST00000061155.12 ENSMUST00000198043.5 ENSMUST00000084952.8 |
Kif9
|
kinesin family member 9 |
| chr2_-_18397547 | 0.33 |
ENSMUST00000091418.12
ENSMUST00000166495.8 |
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr15_+_94441486 | 0.32 |
ENSMUST00000074936.10
|
Irak4
|
interleukin-1 receptor-associated kinase 4 |
| chr5_-_146158235 | 0.32 |
ENSMUST00000161859.8
|
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr15_+_102011352 | 0.32 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr1_-_170042947 | 0.32 |
ENSMUST00000027979.14
ENSMUST00000123399.2 |
Uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr12_+_33004178 | 0.32 |
ENSMUST00000020885.13
|
Sypl
|
synaptophysin-like protein |
| chr15_+_102011415 | 0.32 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr11_-_72686627 | 0.32 |
ENSMUST00000079681.6
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chrX_-_12628309 | 0.31 |
ENSMUST00000096495.11
ENSMUST00000076016.6 |
Med14
|
mediator complex subunit 14 |
| chr18_-_3281089 | 0.31 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr11_-_48707763 | 0.31 |
ENSMUST00000140800.2
|
Trim41
|
tripartite motif-containing 41 |
| chr10_-_39775182 | 0.31 |
ENSMUST00000178045.9
ENSMUST00000178563.3 |
Mfsd4b4
|
major facilitator superfamily domain containing 4B4 |
| chr11_-_4110286 | 0.31 |
ENSMUST00000093381.11
ENSMUST00000101626.9 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr8_+_72973560 | 0.30 |
ENSMUST00000003123.10
|
Fam32a
|
family with sequence similarity 32, member A |
| chr11_-_20062876 | 0.30 |
ENSMUST00000000137.8
|
Actr2
|
ARP2 actin-related protein 2 |
| chr13_-_74956924 | 0.30 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr13_+_41013230 | 0.30 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr11_-_105835238 | 0.30 |
ENSMUST00000019734.11
ENSMUST00000184269.3 ENSMUST00000150563.3 |
Cyb561
|
cytochrome b-561 |
| chr13_-_60325170 | 0.29 |
ENSMUST00000065086.6
|
Gas1
|
growth arrest specific 1 |
| chr8_+_110505184 | 0.29 |
ENSMUST00000093157.13
|
Ap1g1
|
adaptor protein complex AP-1, gamma 1 subunit |
| chr1_+_55170390 | 0.29 |
ENSMUST00000159311.8
ENSMUST00000162364.8 |
Mob4
|
MOB family member 4, phocein |
| chr10_+_34359395 | 0.29 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr11_+_117545037 | 0.29 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr8_+_87350672 | 0.29 |
ENSMUST00000034141.18
ENSMUST00000122188.10 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr4_+_44756553 | 0.28 |
ENSMUST00000107824.9
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr12_-_75642969 | 0.28 |
ENSMUST00000021447.9
ENSMUST00000220035.2 |
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr9_-_88320991 | 0.28 |
ENSMUST00000239462.2
ENSMUST00000165315.9 ENSMUST00000173039.9 |
Snx14
|
sorting nexin 14 |
| chr5_+_117271632 | 0.28 |
ENSMUST00000179276.8
ENSMUST00000092889.12 ENSMUST00000145640.8 |
Taok3
|
TAO kinase 3 |
| chr19_+_6096606 | 0.28 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr13_-_42000958 | 0.27 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr15_-_36609812 | 0.27 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr14_-_20596580 | 0.27 |
ENSMUST00000022355.11
ENSMUST00000161445.8 ENSMUST00000159027.8 |
Ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isoform |
| chr14_+_53980561 | 0.26 |
ENSMUST00000103667.6
|
Trav16
|
T cell receptor alpha variable 16 |
| chr8_+_86567600 | 0.26 |
ENSMUST00000053771.14
ENSMUST00000161850.8 |
Phkb
|
phosphorylase kinase beta |
| chr13_-_95511837 | 0.26 |
ENSMUST00000022189.9
|
Aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr1_+_151631088 | 0.26 |
ENSMUST00000188145.7
ENSMUST00000059498.12 |
Edem3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr4_-_107780716 | 0.26 |
ENSMUST00000106719.8
ENSMUST00000106720.9 ENSMUST00000131644.2 ENSMUST00000030345.15 |
Cpt2
|
carnitine palmitoyltransferase 2 |
| chr1_+_55170476 | 0.25 |
ENSMUST00000162553.2
|
Mob4
|
MOB family member 4, phocein |
| chr4_+_140688514 | 0.25 |
ENSMUST00000010007.9
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr4_+_80828883 | 0.25 |
ENSMUST00000055922.4
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr4_+_44756608 | 0.25 |
ENSMUST00000143385.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr12_-_55045887 | 0.25 |
ENSMUST00000173529.2
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr3_+_137573436 | 0.25 |
ENSMUST00000090178.10
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr5_-_38637624 | 0.25 |
ENSMUST00000067886.12
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr2_-_22930188 | 0.25 |
ENSMUST00000114544.10
ENSMUST00000139038.8 ENSMUST00000126112.8 ENSMUST00000178908.2 ENSMUST00000078977.14 ENSMUST00000140164.8 ENSMUST00000149719.8 |
Abi1
|
abl interactor 1 |
| chr1_+_151631203 | 0.25 |
ENSMUST00000187951.7
ENSMUST00000191070.2 |
Edem3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr10_-_95159933 | 0.25 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr1_-_84817022 | 0.25 |
ENSMUST00000189496.7
ENSMUST00000027421.13 ENSMUST00000186894.7 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr5_-_21990170 | 0.25 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr11_-_44361289 | 0.24 |
ENSMUST00000102795.4
|
Ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr17_-_36290571 | 0.24 |
ENSMUST00000173724.2
ENSMUST00000172900.8 ENSMUST00000174849.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr9_-_50657800 | 0.23 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chrX_+_106860083 | 0.23 |
ENSMUST00000143975.8
ENSMUST00000144695.8 ENSMUST00000167154.2 |
Tent5d
|
terminal nucleotidyltransferase 5D |
| chr19_+_57349112 | 0.23 |
ENSMUST00000036407.6
|
Fam160b1
|
family with sequence similarity 160, member B1 |
| chr15_-_99717956 | 0.23 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr3_+_89958940 | 0.23 |
ENSMUST00000159064.8
|
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr8_-_86567506 | 0.23 |
ENSMUST00000034140.9
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr9_-_88320962 | 0.23 |
ENSMUST00000174806.9
|
Snx14
|
sorting nexin 14 |
| chr6_+_30509826 | 0.23 |
ENSMUST00000031797.11
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr3_-_122778052 | 0.23 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr8_-_81741495 | 0.23 |
ENSMUST00000042724.8
|
Usp38
|
ubiquitin specific peptidase 38 |
| chrX_-_133177717 | 0.23 |
ENSMUST00000087541.12
ENSMUST00000087540.4 |
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr6_+_54016543 | 0.23 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr17_+_85264134 | 0.23 |
ENSMUST00000112305.10
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr6_-_57668992 | 0.22 |
ENSMUST00000053386.6
|
Pyurf
|
Pigy upstream reading frame |
| chrX_+_36059274 | 0.22 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chrX_-_133177638 | 0.22 |
ENSMUST00000113252.8
|
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr6_+_21986445 | 0.22 |
ENSMUST00000115382.8
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr9_-_88320937 | 0.22 |
ENSMUST00000173011.9
|
Snx14
|
sorting nexin 14 |
| chr7_+_101027390 | 0.22 |
ENSMUST00000084895.12
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr9_+_44966464 | 0.22 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chrX_-_72759748 | 0.22 |
ENSMUST00000002091.6
|
Bcap31
|
B cell receptor associated protein 31 |
| chr15_+_88703786 | 0.22 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr13_+_22017906 | 0.22 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr15_-_9529898 | 0.22 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr7_+_143027473 | 0.22 |
ENSMUST00000052348.12
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr13_-_59970383 | 0.22 |
ENSMUST00000225987.2
|
Tut7
|
terminal uridylyl transferase 7 |
| chr9_+_107173907 | 0.22 |
ENSMUST00000168260.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr2_+_166647426 | 0.22 |
ENSMUST00000099078.10
|
Arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr7_+_27770655 | 0.22 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr10_+_34359513 | 0.22 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr17_-_56312555 | 0.21 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr16_+_94171477 | 0.21 |
ENSMUST00000117648.9
ENSMUST00000147352.8 ENSMUST00000150346.8 ENSMUST00000155692.8 ENSMUST00000153988.9 ENSMUST00000139513.9 ENSMUST00000141856.8 ENSMUST00000152117.8 ENSMUST00000150097.8 ENSMUST00000122895.8 ENSMUST00000151770.8 ENSMUST00000231569.2 ENSMUST00000147046.8 ENSMUST00000149885.8 ENSMUST00000127667.8 ENSMUST00000119131.3 ENSMUST00000145883.2 |
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr8_+_110505494 | 0.21 |
ENSMUST00000034171.9
|
Ap1g1
|
adaptor protein complex AP-1, gamma 1 subunit |
| chr19_+_46587523 | 0.21 |
ENSMUST00000138302.9
ENSMUST00000099376.11 |
Wbp1l
|
WW domain binding protein 1 like |
| chr3_-_10505113 | 0.21 |
ENSMUST00000029047.12
ENSMUST00000195822.2 ENSMUST00000099223.11 |
Snx16
|
sorting nexin 16 |
| chr15_-_76079891 | 0.21 |
ENSMUST00000023226.13
|
Plec
|
plectin |
| chr3_-_75464066 | 0.21 |
ENSMUST00000162138.2
ENSMUST00000029424.12 ENSMUST00000161137.8 |
Pdcd10
|
programmed cell death 10 |
| chr9_+_64940004 | 0.21 |
ENSMUST00000167773.2
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr4_+_118266582 | 0.21 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr1_-_43866910 | 0.21 |
ENSMUST00000153317.6
ENSMUST00000128261.2 ENSMUST00000126008.8 ENSMUST00000139451.8 |
Uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr17_-_26063488 | 0.21 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr19_-_10807220 | 0.21 |
ENSMUST00000174176.3
|
Cd6
|
CD6 antigen |
| chr11_-_83193412 | 0.21 |
ENSMUST00000176374.2
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr12_+_11316101 | 0.21 |
ENSMUST00000218866.2
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr16_+_4825216 | 0.21 |
ENSMUST00000185147.8
|
Smim22
|
small integral membrane protein 22 |
| chr3_+_85946145 | 0.20 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr10_+_17598961 | 0.20 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr5_-_69749617 | 0.20 |
ENSMUST00000173927.8
ENSMUST00000120789.8 ENSMUST00000031117.13 |
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr19_+_41017714 | 0.20 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr5_-_146158201 | 0.20 |
ENSMUST00000161574.8
|
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chrX_+_100298134 | 0.20 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chrX_+_106299484 | 0.20 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr9_+_64939695 | 0.20 |
ENSMUST00000034960.14
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr8_-_65302573 | 0.20 |
ENSMUST00000210166.2
|
Klhl2
|
kelch-like 2, Mayven |
| chr7_+_27879650 | 0.20 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr9_-_59260713 | 0.20 |
ENSMUST00000026265.8
|
Bbs4
|
Bardet-Biedl syndrome 4 (human) |
| chr4_+_155666933 | 0.20 |
ENSMUST00000105612.2
|
Nadk
|
NAD kinase |
| chr9_+_65816370 | 0.19 |
ENSMUST00000206594.2
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr13_-_18118736 | 0.19 |
ENSMUST00000009003.9
|
Rala
|
v-ral simian leukemia viral oncogene A (ras related) |
| chr2_-_12424189 | 0.19 |
ENSMUST00000124515.2
|
Mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr9_-_60595401 | 0.19 |
ENSMUST00000114034.9
ENSMUST00000065603.12 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr15_+_58805605 | 0.19 |
ENSMUST00000022980.5
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr3_+_122039206 | 0.19 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr19_-_10807409 | 0.19 |
ENSMUST00000080292.12
|
Cd6
|
CD6 antigen |
| chr3_-_89300936 | 0.19 |
ENSMUST00000124783.8
ENSMUST00000126027.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr8_-_123768759 | 0.19 |
ENSMUST00000098334.13
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr9_+_65816206 | 0.19 |
ENSMUST00000205379.2
ENSMUST00000206048.2 ENSMUST00000034949.10 ENSMUST00000154589.2 |
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr19_-_59932079 | 0.19 |
ENSMUST00000171986.8
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I) |
| chr14_-_20596508 | 0.19 |
ENSMUST00000161989.2
|
Ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isoform |
| chr19_-_10807285 | 0.19 |
ENSMUST00000039043.15
|
Cd6
|
CD6 antigen |
| chr2_+_3425159 | 0.19 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr1_+_55170411 | 0.19 |
ENSMUST00000027122.14
|
Mob4
|
MOB family member 4, phocein |
| chr14_-_26390986 | 0.19 |
ENSMUST00000052932.10
|
Pde12
|
phosphodiesterase 12 |
| chr18_+_30405800 | 0.19 |
ENSMUST00000115812.10
ENSMUST00000115811.8 ENSMUST00000091978.12 |
Pik3c3
|
phosphatidylinositol 3-kinase catalytic subunit type 3 |
| chr6_+_54794433 | 0.18 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chr3_-_79536166 | 0.18 |
ENSMUST00000029386.14
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chr11_-_83189734 | 0.18 |
ENSMUST00000108146.8
ENSMUST00000136369.2 ENSMUST00000018877.9 |
Pex12
|
peroxisomal biogenesis factor 12 |
| chr8_-_65302657 | 0.18 |
ENSMUST00000034017.9
|
Klhl2
|
kelch-like 2, Mayven |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spdef
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 0.8 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 0.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 0.5 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 0.5 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 0.5 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.4 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.9 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.6 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.5 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 1.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.5 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.1 | 0.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.4 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 | 0.4 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.2 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 0.2 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 0.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.2 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.1 | 0.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.8 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.3 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0061354 | chemorepulsion of dopaminergic neuron axon(GO:0036518) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.2 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.7 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.3 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.4 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.6 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.6 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0060807 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) |
| 0.0 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.7 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 1.4 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.4 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:2000169 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.3 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.3 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 0.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.1 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 2.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.5 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.2 | 0.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.7 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.3 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.6 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.3 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.4 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.2 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 0.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0031711 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0051381 | histamine binding(GO:0051381) |
| 0.0 | 0.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |