Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
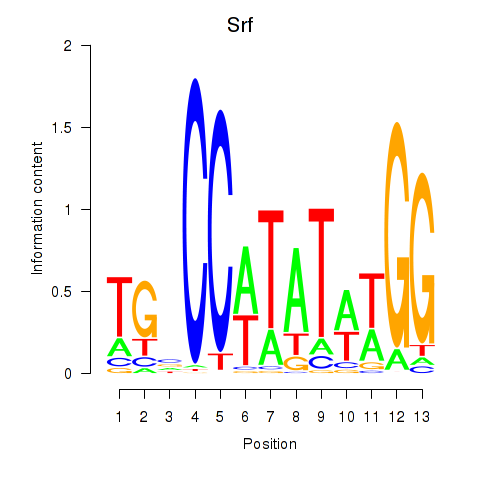
Results for Srf
Z-value: 1.22
Transcription factors associated with Srf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Srf
|
ENSMUSG00000015605.7 | serum response factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srf | mm39_v1_chr17_-_46867083_46867114 | 0.63 | 4.0e-05 | Click! |
Activity profile of Srf motif
Sorted Z-values of Srf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142215027 | 27.22 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr7_-_142215595 | 10.27 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr8_-_124621483 | 9.43 |
ENSMUST00000034453.6
ENSMUST00000212584.2 |
Acta1
|
actin, alpha 1, skeletal muscle |
| chr4_-_119047202 | 8.94 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr10_-_111833138 | 8.85 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr4_+_43957401 | 8.63 |
ENSMUST00000030202.14
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr6_-_83513222 | 7.84 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr4_-_119047167 | 7.50 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_+_43957677 | 7.38 |
ENSMUST00000107855.2
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr2_-_113883285 | 6.46 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr10_+_79824418 | 6.00 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr2_+_156617329 | 5.58 |
ENSMUST00000088552.7
|
Myl9
|
myosin, light polypeptide 9, regulatory |
| chr4_-_119047180 | 5.23 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr11_-_120239339 | 5.21 |
ENSMUST00000071555.13
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr11_-_120239301 | 5.11 |
ENSMUST00000062147.14
ENSMUST00000128055.2 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr6_-_83513184 | 5.08 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr1_-_33796790 | 4.86 |
ENSMUST00000187602.2
ENSMUST00000044691.9 |
Bag2
|
BCL2-associated athanogene 2 |
| chr7_+_3341597 | 4.35 |
ENSMUST00000164553.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr2_+_154390808 | 4.16 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr5_-_100720063 | 4.14 |
ENSMUST00000031264.12
|
Plac8
|
placenta-specific 8 |
| chr2_+_127178072 | 3.95 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr11_-_120238917 | 3.83 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr2_-_152256947 | 3.72 |
ENSMUST00000099207.5
|
Zcchc3
|
zinc finger, CCHC domain containing 3 |
| chr7_+_127846121 | 3.62 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr4_-_132078947 | 3.60 |
ENSMUST00000105951.8
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr9_+_72952115 | 3.53 |
ENSMUST00000184146.8
ENSMUST00000034722.5 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr7_+_127845984 | 3.47 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr7_+_19144950 | 3.20 |
ENSMUST00000208710.2
ENSMUST00000003643.3 |
Ckm
|
creatine kinase, muscle |
| chr2_+_124910037 | 3.14 |
ENSMUST00000070353.4
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr2_+_150628655 | 3.14 |
ENSMUST00000045441.8
|
Pygb
|
brain glycogen phosphorylase |
| chr1_-_43203051 | 3.02 |
ENSMUST00000008280.14
|
Fhl2
|
four and a half LIM domains 2 |
| chr4_-_116228921 | 2.99 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr17_-_57501170 | 2.95 |
ENSMUST00000005976.8
|
Tnfsf14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chrX_+_100492684 | 2.79 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr19_+_53517528 | 2.39 |
ENSMUST00000038287.7
|
Dusp5
|
dual specificity phosphatase 5 |
| chr19_-_5962798 | 2.37 |
ENSMUST00000118623.2
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr19_-_5962862 | 2.35 |
ENSMUST00000136983.8
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr11_-_99383938 | 2.34 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr7_+_101545547 | 2.12 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr3_+_89979948 | 2.10 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr3_+_134918298 | 2.06 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr11_-_5848771 | 1.95 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr18_+_34994253 | 1.91 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr1_+_74448535 | 1.88 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr12_-_103322226 | 1.86 |
ENSMUST00000021617.14
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr5_+_103902020 | 1.82 |
ENSMUST00000054979.10
|
Aff1
|
AF4/FMR2 family, member 1 |
| chr17_+_8144822 | 1.72 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr7_+_101546059 | 1.70 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr17_-_46464441 | 1.69 |
ENSMUST00000171172.3
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr6_-_36787096 | 1.67 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr2_-_13276074 | 1.65 |
ENSMUST00000137670.3
ENSMUST00000114791.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr2_-_13276205 | 1.64 |
ENSMUST00000191959.6
ENSMUST00000028059.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr5_-_134581235 | 1.58 |
ENSMUST00000036999.10
ENSMUST00000100647.7 |
Clip2
|
CAP-GLY domain containing linker protein 2 |
| chr15_-_77726333 | 1.49 |
ENSMUST00000016771.13
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr10_+_4216353 | 1.44 |
ENSMUST00000045730.7
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr2_+_164782642 | 1.43 |
ENSMUST00000137626.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr6_+_42326528 | 1.42 |
ENSMUST00000203329.3
|
Zyx
|
zyxin |
| chr9_+_110592709 | 1.41 |
ENSMUST00000079784.12
|
Myl3
|
myosin, light polypeptide 3 |
| chr6_+_136509922 | 1.39 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr15_-_36794741 | 1.26 |
ENSMUST00000110361.8
ENSMUST00000022894.14 ENSMUST00000110359.2 |
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr7_+_18865610 | 1.18 |
ENSMUST00000165913.2
|
Fbxo46
|
F-box protein 46 |
| chr8_+_15107646 | 1.17 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr15_+_102427149 | 1.14 |
ENSMUST00000146756.8
ENSMUST00000142194.3 |
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chr9_+_75213570 | 1.11 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr16_-_4698148 | 1.10 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr17_+_24022153 | 1.09 |
ENSMUST00000190686.7
ENSMUST00000088621.11 ENSMUST00000233636.2 |
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr12_+_108389075 | 1.08 |
ENSMUST00000109860.8
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr6_+_17306334 | 0.98 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr13_+_23930717 | 0.95 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr7_-_101859379 | 0.92 |
ENSMUST00000210682.2
|
Nup98
|
nucleoporin 98 |
| chr12_+_85520652 | 0.92 |
ENSMUST00000021674.7
|
Fos
|
FBJ osteosarcoma oncogene |
| chr17_+_5991555 | 0.91 |
ENSMUST00000115791.10
ENSMUST00000080283.13 |
Synj2
|
synaptojanin 2 |
| chr14_-_34310602 | 0.91 |
ENSMUST00000064098.14
ENSMUST00000090040.12 ENSMUST00000022330.9 ENSMUST00000022327.13 |
Ldb3
|
LIM domain binding 3 |
| chr4_+_19280850 | 0.85 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr9_-_58220469 | 0.80 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr4_-_141327146 | 0.79 |
ENSMUST00000141518.8
ENSMUST00000127455.8 ENSMUST00000105784.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr14_+_20979466 | 0.74 |
ENSMUST00000022369.9
|
Vcl
|
vinculin |
| chr6_-_113354337 | 0.73 |
ENSMUST00000043333.9
|
Tada3
|
transcriptional adaptor 3 |
| chr1_-_127605660 | 0.71 |
ENSMUST00000160616.8
|
Tmem163
|
transmembrane protein 163 |
| chr7_-_101859308 | 0.67 |
ENSMUST00000070165.7
ENSMUST00000211235.2 ENSMUST00000211022.2 |
Nup98
|
nucleoporin 98 |
| chr14_-_34310637 | 0.66 |
ENSMUST00000227819.2
|
Ldb3
|
LIM domain binding 3 |
| chr18_+_13107535 | 0.65 |
ENSMUST00000234035.2
ENSMUST00000235053.2 |
Impact
|
impact, RWD domain protein |
| chr11_+_101221431 | 0.65 |
ENSMUST00000103105.10
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr6_+_113355076 | 0.65 |
ENSMUST00000156898.5
ENSMUST00000203578.3 ENSMUST00000171058.8 |
Arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr5_-_148336574 | 0.62 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr15_+_84926909 | 0.52 |
ENSMUST00000229203.2
|
Fam118a
|
family with sequence similarity 118, member A |
| chr5_-_113044216 | 0.50 |
ENSMUST00000086617.11
|
Myo18b
|
myosin XVIIIb |
| chr10_+_32959472 | 0.43 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr3_+_106020545 | 0.42 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr17_-_46867083 | 0.40 |
ENSMUST00000015749.7
|
Srf
|
serum response factor |
| chr16_+_57369595 | 0.38 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr11_+_82802079 | 0.36 |
ENSMUST00000018989.14
ENSMUST00000164945.3 |
Unc45b
|
unc-45 myosin chaperone B |
| chr6_-_97156032 | 0.31 |
ENSMUST00000095664.6
|
Tmf1
|
TATA element modulatory factor 1 |
| chr4_-_122779837 | 0.30 |
ENSMUST00000106255.8
ENSMUST00000106257.10 |
Cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr2_-_127363251 | 0.29 |
ENSMUST00000028850.15
ENSMUST00000103215.11 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr19_+_12655487 | 0.25 |
ENSMUST00000215134.2
ENSMUST00000049724.8 |
Olfr1443
|
olfactory receptor 1443 |
| chr17_+_28259749 | 0.24 |
ENSMUST00000233869.2
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr15_+_39255185 | 0.22 |
ENSMUST00000228839.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr9_+_74959259 | 0.22 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr10_+_90412827 | 0.21 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_-_43025792 | 0.17 |
ENSMUST00000067481.6
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr7_-_101859033 | 0.16 |
ENSMUST00000211005.2
|
Nup98
|
nucleoporin 98 |
| chr6_-_113354468 | 0.15 |
ENSMUST00000099118.8
|
Tada3
|
transcriptional adaptor 3 |
| chr6_+_87350292 | 0.15 |
ENSMUST00000032128.6
|
Gkn2
|
gastrokine 2 |
| chr4_-_43025756 | 0.14 |
ENSMUST00000098109.9
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr14_-_34310438 | 0.14 |
ENSMUST00000228044.2
ENSMUST00000022328.14 |
Ldb3
|
LIM domain binding 3 |
| chr6_-_68840015 | 0.13 |
ENSMUST00000103336.2
|
Igkv1-88
|
immunoglobulin kappa chain variable 1-88 |
| chr7_-_119319965 | 0.12 |
ENSMUST00000033236.9
|
Thumpd1
|
THUMP domain containing 1 |
| chr11_+_19874354 | 0.10 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr13_-_103094784 | 0.10 |
ENSMUST00000172264.8
ENSMUST00000099202.10 |
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr13_+_109397184 | 0.08 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr3_+_68375495 | 0.07 |
ENSMUST00000182532.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr15_+_11000802 | 0.03 |
ENSMUST00000117100.4
|
Slc45a2
|
solute carrier family 45, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Srf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 37.5 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 1.6 | 15.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 1.2 | 3.5 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 1.0 | 2.9 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.8 | 3.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.7 | 7.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.6 | 1.9 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.5 | 4.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 2.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.5 | 1.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.5 | 1.9 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.4 | 1.7 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.4 | 1.5 | GO:1903921 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.4 | 1.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 2.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 3.8 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 6.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.2 | 3.0 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 1.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 14.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 1.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 15.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 0.5 | GO:0072755 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.2 | 1.0 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 2.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.6 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.1 | 4.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.3 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 1.7 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 3.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 3.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.4 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 6.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 1.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 1.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.9 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.9 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 3.3 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 3.2 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 | 0.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 4.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) positive regulation of inhibitory postsynaptic potential(GO:0097151) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.4 | 3.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 1.8 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 1.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 14.9 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 11.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 0.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 5.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 2.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 3.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 5.4 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 1.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 1.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.9 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 16.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 9.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 6.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 42.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 9.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 17.1 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.8 | 37.5 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.8 | 3.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 3.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 3.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.5 | 3.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 4.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 1.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 6.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 2.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.9 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.3 | 5.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 0.7 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 1.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 1.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.9 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 10.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 2.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 13.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.9 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 1.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.4 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 2.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 5.1 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 27.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 37.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 11.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 6.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 8.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 3.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 37.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 21.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 3.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 9.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.6 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 1.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 2.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 3.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |