Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
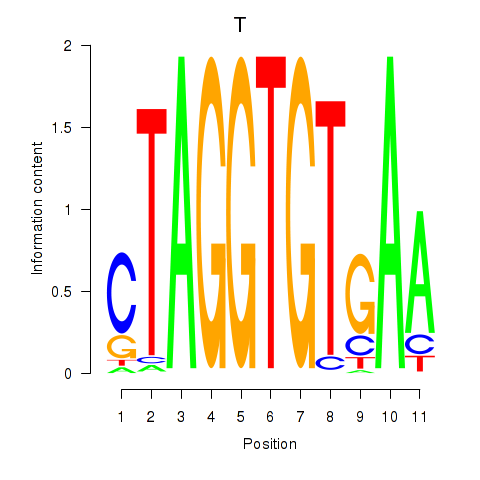
Results for T
Z-value: 0.68
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSMUSG00000062327.11 | brachyury, T-box transcription factor T |
Activity profile of T motif
Sorted Z-values of T motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_20579322 | 3.99 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr8_-_106660470 | 3.96 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr15_+_9335636 | 3.08 |
ENSMUST00000072403.7
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr1_+_88034556 | 2.55 |
ENSMUST00000113137.2
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr1_+_171246593 | 2.07 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chrX_+_138511360 | 1.87 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr19_-_20704896 | 1.81 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr11_-_94283792 | 1.64 |
ENSMUST00000178136.8
ENSMUST00000021231.8 |
Abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr1_+_88066086 | 1.61 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr6_+_90310252 | 1.59 |
ENSMUST00000046128.12
ENSMUST00000164761.6 |
Uroc1
|
urocanase domain containing 1 |
| chr9_-_78263025 | 1.53 |
ENSMUST00000125479.8
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr15_-_82291372 | 1.53 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr9_+_37524966 | 1.46 |
ENSMUST00000215474.2
|
Siae
|
sialic acid acetylesterase |
| chr10_-_95678786 | 1.36 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr10_-_95678748 | 1.34 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr1_-_93373145 | 1.29 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr2_-_147888816 | 1.28 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr2_-_27138347 | 1.24 |
ENSMUST00000139312.8
|
Sardh
|
sarcosine dehydrogenase |
| chr8_-_70664901 | 1.21 |
ENSMUST00000063788.8
ENSMUST00000110127.8 |
Slc25a42
|
solute carrier family 25, member 42 |
| chr9_-_9239056 | 1.20 |
ENSMUST00000093893.12
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr6_+_40619913 | 1.17 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chr2_+_154975419 | 1.16 |
ENSMUST00000029126.15
ENSMUST00000109685.8 |
Itch
|
itchy, E3 ubiquitin protein ligase |
| chr7_-_140462187 | 1.16 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr7_-_140462221 | 1.15 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr18_-_12369351 | 1.10 |
ENSMUST00000025279.6
|
Npc1
|
NPC intracellular cholesterol transporter 1 |
| chr7_-_140461769 | 1.07 |
ENSMUST00000106048.10
ENSMUST00000147331.9 ENSMUST00000137710.2 |
Sirt3
|
sirtuin 3 |
| chr16_+_93404719 | 1.05 |
ENSMUST00000039659.9
ENSMUST00000231762.2 |
Cbr1
|
carbonyl reductase 1 |
| chr14_+_6226418 | 1.05 |
ENSMUST00000112625.9
|
Oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr5_-_38649291 | 1.01 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr2_-_51862941 | 1.00 |
ENSMUST00000145481.8
ENSMUST00000112705.9 |
Nmi
|
N-myc (and STAT) interactor |
| chr4_+_155648256 | 0.96 |
ENSMUST00000143840.2
ENSMUST00000146080.8 |
Nadk
|
NAD kinase |
| chr3_-_121076745 | 0.95 |
ENSMUST00000135818.8
ENSMUST00000137234.2 |
Tlcd4
|
TLC domain containing 4 |
| chr16_-_18232202 | 0.95 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr2_+_32498997 | 0.93 |
ENSMUST00000143625.2
ENSMUST00000128811.2 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr15_-_60793115 | 0.92 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr1_-_93373364 | 0.90 |
ENSMUST00000190321.7
ENSMUST00000042498.14 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr19_+_10160249 | 0.89 |
ENSMUST00000010807.6
|
Fads1
|
fatty acid desaturase 1 |
| chr10_-_34294461 | 0.88 |
ENSMUST00000213269.2
ENSMUST00000099973.4 ENSMUST00000105512.8 ENSMUST00000047885.14 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr9_-_43151179 | 0.87 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr2_-_103627937 | 0.86 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1 |
| chrX_-_161612373 | 0.83 |
ENSMUST00000041370.11
ENSMUST00000112316.9 ENSMUST00000112315.2 |
Txlng
|
taxilin gamma |
| chr1_+_132243849 | 0.82 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr9_+_13677266 | 0.81 |
ENSMUST00000152532.8
|
Mtmr2
|
myotubularin related protein 2 |
| chr19_-_32080496 | 0.81 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr5_-_108280350 | 0.79 |
ENSMUST00000119784.2
ENSMUST00000117759.2 |
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr17_-_78991691 | 0.77 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr16_+_27207722 | 0.77 |
ENSMUST00000039443.14
ENSMUST00000096127.11 |
Ccdc50
|
coiled-coil domain containing 50 |
| chr4_-_12087911 | 0.77 |
ENSMUST00000050686.10
|
Tmem67
|
transmembrane protein 67 |
| chr18_+_38429792 | 0.73 |
ENSMUST00000237211.2
|
Rnf14
|
ring finger protein 14 |
| chr9_-_53521585 | 0.72 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr14_+_55120777 | 0.71 |
ENSMUST00000022806.10
|
Bcl2l2
|
BCL2-like 2 |
| chr3_-_107851021 | 0.70 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr9_-_57347366 | 0.70 |
ENSMUST00000214144.2
ENSMUST00000085709.6 ENSMUST00000214624.2 ENSMUST00000215883.2 ENSMUST00000214339.2 ENSMUST00000215299.2 ENSMUST00000214166.2 ENSMUST00000214065.2 |
Ppcdc
|
phosphopantothenoylcysteine decarboxylase |
| chr11_+_29476385 | 0.69 |
ENSMUST00000133452.8
|
Mtif2
|
mitochondrial translational initiation factor 2 |
| chr6_-_94677118 | 0.69 |
ENSMUST00000101126.3
ENSMUST00000032105.11 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr3_+_152052102 | 0.67 |
ENSMUST00000117492.9
ENSMUST00000026507.13 ENSMUST00000197748.5 |
Usp33
|
ubiquitin specific peptidase 33 |
| chr7_-_71956332 | 0.66 |
ENSMUST00000079323.8
|
Mctp2
|
multiple C2 domains, transmembrane 2 |
| chr6_-_54941673 | 0.65 |
ENSMUST00000203837.2
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr7_-_79492091 | 0.64 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr3_+_40905066 | 0.64 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr8_+_107662352 | 0.63 |
ENSMUST00000212524.2
ENSMUST00000047425.5 |
Sntb2
|
syntrophin, basic 2 |
| chr6_+_52691204 | 0.61 |
ENSMUST00000138040.8
ENSMUST00000129660.2 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr14_+_55120875 | 0.61 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr3_+_40905216 | 0.61 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr1_+_44158111 | 0.61 |
ENSMUST00000155917.8
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr19_+_37686240 | 0.60 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr9_+_86349226 | 0.58 |
ENSMUST00000188675.7
|
Dop1a
|
DOP1 leucine zipper like protein A |
| chr6_+_48624295 | 0.54 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr11_+_93935021 | 0.54 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr19_+_28941292 | 0.52 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr9_+_86349207 | 0.52 |
ENSMUST00000190957.7
|
Dop1a
|
DOP1 leucine zipper like protein A |
| chr4_-_129121676 | 0.52 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr6_-_116084810 | 0.52 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr15_+_95688763 | 0.51 |
ENSMUST00000227791.2
|
Ano6
|
anoctamin 6 |
| chr5_+_24633206 | 0.49 |
ENSMUST00000115049.9
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr18_+_38429688 | 0.48 |
ENSMUST00000170811.8
ENSMUST00000072376.13 ENSMUST00000237903.2 ENSMUST00000236116.2 ENSMUST00000237824.2 ENSMUST00000236982.2 ENSMUST00000235549.2 ENSMUST00000237416.2 ENSMUST00000237089.2 |
Rnf14
|
ring finger protein 14 |
| chr11_+_87938626 | 0.47 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr6_+_125090473 | 0.46 |
ENSMUST00000124317.2
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr10_+_21854540 | 0.46 |
ENSMUST00000142174.8
ENSMUST00000164659.8 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr9_-_20657643 | 0.46 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chr6_-_71376277 | 0.46 |
ENSMUST00000149415.2
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr2_-_51863203 | 0.44 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr9_+_110948492 | 0.44 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr11_+_93935156 | 0.43 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr7_-_89166781 | 0.42 |
ENSMUST00000041761.7
|
Prss23
|
protease, serine 23 |
| chr11_-_78427061 | 0.42 |
ENSMUST00000017759.9
ENSMUST00000108277.3 |
Tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr1_-_155908695 | 0.41 |
ENSMUST00000136397.8
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr5_-_31250817 | 0.41 |
ENSMUST00000031037.14
|
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr4_-_129132963 | 0.41 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr9_+_50405817 | 0.41 |
ENSMUST00000114474.8
ENSMUST00000188047.2 |
Plet1
|
placenta expressed transcript 1 |
| chr16_+_84631956 | 0.40 |
ENSMUST00000009120.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr16_+_84631789 | 0.40 |
ENSMUST00000114184.8
|
Gabpa
|
GA repeat binding protein, alpha |
| chr11_+_93935066 | 0.39 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr3_+_90138895 | 0.38 |
ENSMUST00000029546.15
ENSMUST00000119304.2 |
Jtb
|
jumping translocation breakpoint |
| chr9_-_95727267 | 0.37 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform) |
| chr2_-_180844582 | 0.37 |
ENSMUST00000016511.6
|
Ptk6
|
PTK6 protein tyrosine kinase 6 |
| chr1_-_16689660 | 0.37 |
ENSMUST00000117146.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr18_+_38429858 | 0.36 |
ENSMUST00000171461.3
ENSMUST00000235811.2 |
Rnf14
|
ring finger protein 14 |
| chr3_+_144998233 | 0.36 |
ENSMUST00000029848.5
ENSMUST00000139001.2 |
Col24a1
|
collagen, type XXIV, alpha 1 |
| chr11_+_49094292 | 0.35 |
ENSMUST00000150284.8
ENSMUST00000109197.8 ENSMUST00000151228.2 |
Zfp62
|
zinc finger protein 62 |
| chr11_+_93934940 | 0.34 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr11_+_79551358 | 0.34 |
ENSMUST00000155381.2
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr7_-_89166724 | 0.29 |
ENSMUST00000208888.2
|
Prss23
|
protease, serine 23 |
| chr16_-_44566700 | 0.29 |
ENSMUST00000023348.11
ENSMUST00000162512.8 |
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr15_-_79025387 | 0.28 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chr6_-_69800923 | 0.28 |
ENSMUST00000103368.3
|
Igkv5-43
|
immunoglobulin kappa chain variable 5-43 |
| chr6_-_116693849 | 0.26 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr4_+_53440389 | 0.26 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr10_+_88295515 | 0.25 |
ENSMUST00000125612.2
|
Sycp3
|
synaptonemal complex protein 3 |
| chr15_-_59245998 | 0.25 |
ENSMUST00000022976.6
|
Washc5
|
WASH complex subunit 5 |
| chr11_-_69556888 | 0.24 |
ENSMUST00000108654.3
|
Cd68
|
CD68 antigen |
| chr9_-_110818679 | 0.23 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr10_+_23672842 | 0.20 |
ENSMUST00000119597.8
ENSMUST00000179321.8 ENSMUST00000133289.2 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr6_+_48624158 | 0.20 |
ENSMUST00000203083.3
|
Gimap8
|
GTPase, IMAP family member 8 |
| chr11_+_78427181 | 0.20 |
ENSMUST00000128788.8
|
Ift20
|
intraflagellar transport 20 |
| chr7_-_30826184 | 0.20 |
ENSMUST00000211945.2
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr15_-_76004395 | 0.19 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr11_-_69556904 | 0.19 |
ENSMUST00000018918.12
|
Cd68
|
CD68 antigen |
| chr11_+_49094119 | 0.19 |
ENSMUST00000109198.8
ENSMUST00000137061.9 |
Zfp62
|
zinc finger protein 62 |
| chr8_-_73246435 | 0.19 |
ENSMUST00000152080.8
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr4_+_116453927 | 0.19 |
ENSMUST00000051869.8
|
Ccdc17
|
coiled-coil domain containing 17 |
| chr7_-_6733411 | 0.18 |
ENSMUST00000239104.2
ENSMUST00000051209.11 |
Peg3
|
paternally expressed 3 |
| chr9_-_119766580 | 0.18 |
ENSMUST00000035099.9
|
Gorasp1
|
golgi reassembly stacking protein 1 |
| chr2_+_32340459 | 0.18 |
ENSMUST00000048431.3
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr11_+_87938128 | 0.17 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr10_+_128158328 | 0.17 |
ENSMUST00000219037.2
ENSMUST00000026446.4 |
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr1_-_156767196 | 0.17 |
ENSMUST00000185198.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr15_+_95688712 | 0.16 |
ENSMUST00000071874.8
|
Ano6
|
anoctamin 6 |
| chr4_-_3574844 | 0.16 |
ENSMUST00000029891.12
|
Tmem68
|
transmembrane protein 68 |
| chr11_+_78427219 | 0.16 |
ENSMUST00000050366.15
ENSMUST00000108275.2 |
Ift20
|
intraflagellar transport 20 |
| chr11_-_98084086 | 0.16 |
ENSMUST00000092735.12
ENSMUST00000107545.9 |
Med1
|
mediator complex subunit 1 |
| chr1_-_16689527 | 0.15 |
ENSMUST00000182554.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr8_+_70107131 | 0.15 |
ENSMUST00000204285.3
|
Zfp964
|
zinc finger protein 964 |
| chr8_-_34333573 | 0.14 |
ENSMUST00000183062.2
|
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr8_+_106363141 | 0.14 |
ENSMUST00000005841.16
|
Ctcf
|
CCCTC-binding factor |
| chr9_-_56151334 | 0.14 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_-_86435579 | 0.13 |
ENSMUST00000138810.3
ENSMUST00000058286.9 ENSMUST00000154617.8 |
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1 |
| chr7_+_43093507 | 0.13 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr6_+_68026941 | 0.12 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr3_-_126918491 | 0.12 |
ENSMUST00000238781.2
|
Ank2
|
ankyrin 2, brain |
| chr15_-_81244940 | 0.11 |
ENSMUST00000023040.9
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr16_-_44566641 | 0.10 |
ENSMUST00000161436.2
|
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr17_-_36206812 | 0.10 |
ENSMUST00000059740.15
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr6_-_119444157 | 0.09 |
ENSMUST00000118120.8
|
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr9_-_42368880 | 0.09 |
ENSMUST00000125995.8
|
Tbcel
|
tubulin folding cofactor E-like |
| chr11_+_105069591 | 0.09 |
ENSMUST00000106939.9
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr11_+_87938519 | 0.09 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr9_-_40442669 | 0.09 |
ENSMUST00000119373.9
|
Gramd1b
|
GRAM domain containing 1B |
| chr17_-_25564501 | 0.08 |
ENSMUST00000153118.2
ENSMUST00000146856.3 |
Tpsab1
|
tryptase alpha/beta 1 |
| chr14_-_24294933 | 0.08 |
ENSMUST00000169880.3
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr2_-_125993887 | 0.08 |
ENSMUST00000110448.3
ENSMUST00000110446.9 |
Fam227b
|
family with sequence similarity 227, member B |
| chrX_-_138683102 | 0.07 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr14_+_13803477 | 0.06 |
ENSMUST00000102996.4
|
Cfap20dc
|
CFAP20 domain containing |
| chr5_+_129924564 | 0.06 |
ENSMUST00000041466.14
|
Zbed5
|
zinc finger, BED type containing 5 |
| chr12_-_75596441 | 0.06 |
ENSMUST00000218716.2
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B', epsilon |
| chr17_+_84819260 | 0.05 |
ENSMUST00000047206.7
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr2_+_181134977 | 0.04 |
ENSMUST00000069649.9
|
Abhd16b
|
abhydrolase domain containing 16B |
| chr10_+_88295431 | 0.04 |
ENSMUST00000020252.10
|
Sycp3
|
synaptonemal complex protein 3 |
| chr12_+_84363603 | 0.03 |
ENSMUST00000045931.12
|
Zfp410
|
zinc finger protein 410 |
| chr13_+_104315301 | 0.02 |
ENSMUST00000022225.12
ENSMUST00000069187.12 |
Trim23
|
tripartite motif-containing 23 |
| chr16_-_44153498 | 0.02 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr7_-_119694400 | 0.02 |
ENSMUST00000209154.3
ENSMUST00000046993.4 |
Dnah3
|
dynein, axonemal, heavy chain 3 |
| chr12_-_113625906 | 0.01 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr7_+_5023552 | 0.00 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
Network of associatons between targets according to the STRING database.
First level regulatory network of T
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.4 | 4.0 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 1.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 1.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.4 | 1.2 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) nucleoside transmembrane transport(GO:1901642) |
| 0.3 | 4.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.6 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.3 | 1.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 3.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 0.7 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 1.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 1.0 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.2 | 1.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.7 | GO:0097045 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.2 | 1.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 1.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 1.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 2.4 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 4.6 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:0051878 | lateral element assembly(GO:0051878) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 1.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.8 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.4 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 1.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.7 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.2 | GO:0070318 | enucleate erythrocyte development(GO:0048822) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 1.9 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.3 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.4 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.4 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 1.6 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.6 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 1.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.5 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.7 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 2.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.3 | GO:0000802 | transverse filament(GO:0000802) |
| 0.1 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 5.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.4 | 1.2 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.3 | 1.0 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 0.9 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.3 | 1.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 1.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 1.2 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.2 | 1.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 3.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 7.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.7 | GO:0048273 | MAP-kinase scaffold activity(GO:0005078) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 1.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.5 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 1.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 1.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |