Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
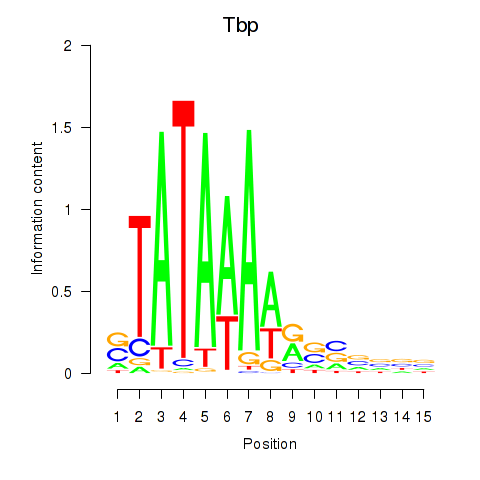
Results for Tbp
Z-value: 3.50
Transcription factors associated with Tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbp
|
ENSMUSG00000014767.18 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbp | mm39_v1_chr17_+_15720150_15720222 | 0.53 | 9.3e-04 | Click! |
Activity profile of Tbp motif
Sorted Z-values of Tbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_90603013 | 42.90 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr11_+_87684299 | 33.63 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr5_+_90920294 | 29.10 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr5_+_90920353 | 27.87 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chr9_+_98372575 | 27.82 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr4_+_120523758 | 25.96 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr7_-_103463120 | 25.60 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr6_-_41291634 | 23.71 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr17_-_31348576 | 23.42 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr7_-_103477126 | 22.12 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr7_-_45175570 | 21.46 |
ENSMUST00000167273.2
ENSMUST00000042105.11 |
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr8_+_94905710 | 21.43 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr6_-_41354538 | 21.21 |
ENSMUST00000096003.7
|
Prss3
|
protease, serine 3 |
| chr1_+_134110142 | 21.20 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr14_-_56339915 | 20.57 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr7_-_4815111 | 19.84 |
ENSMUST00000205885.2
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr3_-_20329823 | 17.75 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr11_-_55075855 | 17.60 |
ENSMUST00000039305.6
|
Slc36a2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr7_-_4815542 | 17.45 |
ENSMUST00000079496.9
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr8_+_85696216 | 16.95 |
ENSMUST00000109734.8
ENSMUST00000005292.15 |
Prdx2
|
peroxiredoxin 2 |
| chr1_+_134109888 | 16.20 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr8_+_85696453 | 16.02 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr11_-_99328969 | 15.65 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr6_+_41331039 | 15.13 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr7_-_143056252 | 15.03 |
ENSMUST00000010904.5
|
Phlda2
|
pleckstrin homology like domain, family A, member 2 |
| chr8_+_85696396 | 14.90 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr9_+_7558449 | 14.36 |
ENSMUST00000018765.4
|
Mmp8
|
matrix metallopeptidase 8 |
| chr2_-_164197987 | 13.75 |
ENSMUST00000165980.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr13_-_22017677 | 13.73 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr6_-_41012435 | 13.65 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr11_-_98915005 | 13.64 |
ENSMUST00000068031.8
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr6_-_87958611 | 13.60 |
ENSMUST00000056403.7
|
H1f10
|
H1.10 linker histone |
| chr6_-_57802131 | 13.57 |
ENSMUST00000204878.3
ENSMUST00000145608.7 ENSMUST00000203212.3 ENSMUST00000114297.5 |
Vopp1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr2_-_164198427 | 13.50 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr6_-_72935468 | 13.50 |
ENSMUST00000114050.8
|
Tmsb10
|
thymosin, beta 10 |
| chr6_+_41435846 | 13.00 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chr11_-_69838971 | 12.65 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr6_+_41369290 | 12.56 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chr11_-_102787950 | 12.33 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr7_-_44181477 | 12.18 |
ENSMUST00000098483.9
ENSMUST00000035323.6 |
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr8_+_85696695 | 11.78 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr6_-_72935382 | 11.63 |
ENSMUST00000144337.2
|
Tmsb10
|
thymosin, beta 10 |
| chr11_-_115405200 | 11.58 |
ENSMUST00000021083.7
|
Jpt1
|
Jupiter microtubule associated homolog 1 |
| chr2_+_119449192 | 11.55 |
ENSMUST00000028771.8
|
Nusap1
|
nucleolar and spindle associated protein 1 |
| chr18_-_78166595 | 11.46 |
ENSMUST00000091813.12
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr7_-_103502404 | 11.40 |
ENSMUST00000033229.5
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr4_-_41098174 | 11.35 |
ENSMUST00000055327.8
|
Aqp3
|
aquaporin 3 |
| chr17_+_24939072 | 11.15 |
ENSMUST00000054289.13
|
Rps2
|
ribosomal protein S2 |
| chr17_+_24939225 | 10.27 |
ENSMUST00000146867.2
|
Rps2
|
ribosomal protein S2 |
| chr11_+_76889415 | 10.04 |
ENSMUST00000108402.9
ENSMUST00000021195.11 |
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr17_+_24939037 | 10.01 |
ENSMUST00000170715.8
|
Rps2
|
ribosomal protein S2 |
| chr13_-_22225527 | 9.90 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr2_+_13579092 | 9.88 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr18_-_78166539 | 9.83 |
ENSMUST00000160292.8
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr2_+_13578738 | 9.50 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr7_-_126651847 | 9.50 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr17_-_34043502 | 9.50 |
ENSMUST00000087342.13
ENSMUST00000173844.8 |
Rps28
|
ribosomal protein S28 |
| chr6_-_72935171 | 9.21 |
ENSMUST00000114049.2
|
Tmsb10
|
thymosin, beta 10 |
| chr18_+_34758062 | 9.15 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr17_-_34043320 | 8.98 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr17_+_23945310 | 8.84 |
ENSMUST00000024701.9
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr1_-_134006847 | 8.81 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr2_-_119448935 | 8.72 |
ENSMUST00000123818.2
|
Oip5
|
Opa interacting protein 5 |
| chr8_-_106660470 | 8.59 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr4_-_132080916 | 8.32 |
ENSMUST00000155129.2
ENSMUST00000151374.2 |
Rcc1
Snhg3
|
regulator of chromosome condensation 1 small nucleolar RNA host gene 3 |
| chr15_-_79967543 | 8.32 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr18_+_34757687 | 7.91 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chr2_-_113883285 | 7.91 |
ENSMUST00000090269.7
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr10_-_35587888 | 7.87 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr10_-_128462616 | 7.80 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr11_+_94827050 | 7.66 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr7_-_15993110 | 7.57 |
ENSMUST00000168818.2
|
C5ar1
|
complement component 5a receptor 1 |
| chr9_-_61821820 | 7.55 |
ENSMUST00000008036.9
|
Rplp1
|
ribosomal protein, large, P1 |
| chr4_+_118965908 | 7.51 |
ENSMUST00000030398.10
|
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr18_+_34757666 | 7.40 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr15_-_66841465 | 7.25 |
ENSMUST00000170903.8
ENSMUST00000166420.8 ENSMUST00000005256.14 ENSMUST00000164070.2 |
Ndrg1
|
N-myc downstream regulated gene 1 |
| chr10_-_80512117 | 7.21 |
ENSMUST00000200082.5
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr11_-_101998648 | 7.14 |
ENSMUST00000177304.8
ENSMUST00000017455.15 |
Pyy
|
peptide YY |
| chr11_+_67167950 | 6.97 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr17_-_34174631 | 6.87 |
ENSMUST00000174609.9
ENSMUST00000008812.9 |
Rps18
|
ribosomal protein S18 |
| chr2_-_25114660 | 6.80 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr4_+_118965992 | 6.75 |
ENSMUST00000134105.8
ENSMUST00000144329.8 ENSMUST00000208090.2 |
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr1_-_172722589 | 6.73 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr4_-_3835595 | 6.65 |
ENSMUST00000138502.2
|
Rps20
|
ribosomal protein S20 |
| chr17_-_80514725 | 6.35 |
ENSMUST00000234696.2
ENSMUST00000235069.2 ENSMUST00000063417.11 |
Srsf7
|
serine and arginine-rich splicing factor 7 |
| chr6_-_40976413 | 6.32 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr11_-_33113071 | 6.31 |
ENSMUST00000093201.13
ENSMUST00000101375.5 ENSMUST00000109354.10 ENSMUST00000075641.10 |
Npm1
|
nucleophosmin 1 |
| chr2_+_152578164 | 6.29 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr13_+_21900554 | 6.10 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr5_+_139239247 | 6.09 |
ENSMUST00000138508.8
ENSMUST00000110878.2 |
Get4
|
golgi to ER traffic protein 4 |
| chr6_+_4747298 | 5.97 |
ENSMUST00000166678.2
ENSMUST00000176204.8 |
Peg10
|
paternally expressed 10 |
| chr6_+_78402956 | 5.83 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr4_+_126915104 | 5.75 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr4_+_34893772 | 5.64 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr3_+_145855929 | 5.60 |
ENSMUST00000098524.5
|
Mcoln2
|
mucolipin 2 |
| chr9_+_108385247 | 5.59 |
ENSMUST00000207810.2
ENSMUST00000207862.2 ENSMUST00000208581.2 ENSMUST00000134939.9 ENSMUST00000207713.2 |
Qars
|
glutaminyl-tRNA synthetase |
| chr3_-_20296337 | 5.52 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr10_-_88440869 | 5.47 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr11_-_102787972 | 5.43 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chr2_+_122479770 | 5.28 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr8_-_71292295 | 5.24 |
ENSMUST00000212405.2
ENSMUST00000002989.11 |
Arrdc2
|
arrestin domain containing 2 |
| chr2_+_90927053 | 5.14 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr7_-_30643444 | 5.12 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr10_-_88440996 | 5.09 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr6_+_40941688 | 5.09 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr2_+_36120438 | 5.01 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr1_-_66974492 | 5.00 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr3_+_142470806 | 4.92 |
ENSMUST00000029938.10
|
Gtf2b
|
general transcription factor IIB |
| chr13_+_21901791 | 4.84 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr5_+_142946098 | 4.83 |
ENSMUST00000031565.15
ENSMUST00000198017.5 |
Fscn1
|
fascin actin-bundling protein 1 |
| chr6_-_4747157 | 4.82 |
ENSMUST00000126151.8
ENSMUST00000115577.9 ENSMUST00000101677.9 ENSMUST00000115579.8 ENSMUST00000004750.15 |
Sgce
|
sarcoglycan, epsilon |
| chr19_+_39049442 | 4.82 |
ENSMUST00000087236.5
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr7_+_130633776 | 4.79 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr4_-_116484675 | 4.78 |
ENSMUST00000081182.5
ENSMUST00000030457.12 |
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr17_+_29251602 | 4.76 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr1_-_125362321 | 4.73 |
ENSMUST00000191544.7
|
Actr3
|
ARP3 actin-related protein 3 |
| chr18_+_36661198 | 4.72 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr6_-_87649173 | 4.66 |
ENSMUST00000032130.8
|
Aplf
|
aprataxin and PNKP like factor |
| chr10_-_40178182 | 4.64 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr12_+_102521225 | 4.59 |
ENSMUST00000021610.7
|
Chga
|
chromogranin A |
| chr6_-_112673565 | 4.57 |
ENSMUST00000113182.8
ENSMUST00000113180.8 ENSMUST00000068487.12 ENSMUST00000077088.11 |
Rad18
|
RAD18 E3 ubiquitin protein ligase |
| chrX_+_73314418 | 4.57 |
ENSMUST00000008826.14
ENSMUST00000151702.8 ENSMUST00000074085.12 ENSMUST00000135690.2 |
Rpl10
|
ribosomal protein L10 |
| chr6_-_87649124 | 4.49 |
ENSMUST00000065997.5
|
Aplf
|
aprataxin and PNKP like factor |
| chr4_+_63478478 | 4.48 |
ENSMUST00000080336.4
|
Tmem268
|
transmembrane protein 268 |
| chr5_+_33176160 | 4.43 |
ENSMUST00000019109.8
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr11_+_85202990 | 4.43 |
ENSMUST00000127717.2
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr17_-_34000804 | 4.40 |
ENSMUST00000002360.17
|
Angptl4
|
angiopoietin-like 4 |
| chr11_-_100012384 | 4.40 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr9_-_67739607 | 4.35 |
ENSMUST00000054500.7
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr1_-_154975376 | 4.33 |
ENSMUST00000055322.6
|
Ier5
|
immediate early response 5 |
| chr2_-_62313981 | 4.29 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr6_-_34294377 | 4.19 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr13_-_23946359 | 4.10 |
ENSMUST00000091701.3
|
H3c1
|
H3 clustered histone 1 |
| chr13_+_23947641 | 4.08 |
ENSMUST00000055770.4
|
H1f1
|
H1.1 linker histone, cluster member |
| chr19_+_39102342 | 4.03 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr13_-_21967540 | 3.98 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11 |
| chr8_-_85696040 | 3.96 |
ENSMUST00000214133.2
ENSMUST00000147812.8 |
Gm49661
Rnaseh2a
|
predicted gene, 49661 ribonuclease H2, large subunit |
| chr10_+_79613083 | 3.88 |
ENSMUST00000020575.5
|
Fstl3
|
follistatin-like 3 |
| chr19_-_3464447 | 3.83 |
ENSMUST00000025842.8
ENSMUST00000237521.2 |
Gal
|
galanin and GMAP prepropeptide |
| chr4_-_119515978 | 3.78 |
ENSMUST00000106309.9
ENSMUST00000044426.8 |
Guca2b
|
guanylate cyclase activator 2b (retina) |
| chr13_+_95460110 | 3.78 |
ENSMUST00000221807.2
|
Zbed3
|
zinc finger, BED type containing 3 |
| chr6_-_72416531 | 3.76 |
ENSMUST00000205335.2
ENSMUST00000206692.2 ENSMUST00000059472.10 |
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr11_+_62442502 | 3.66 |
ENSMUST00000136938.2
|
Ubb
|
ubiquitin B |
| chr4_-_116485118 | 3.56 |
ENSMUST00000030456.14
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr13_-_23929490 | 3.44 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr14_+_69846517 | 3.36 |
ENSMUST00000022660.14
|
Loxl2
|
lysyl oxidase-like 2 |
| chr8_+_108162985 | 3.32 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_-_100026754 | 3.32 |
ENSMUST00000107411.3
|
Krt15
|
keratin 15 |
| chr14_-_57015748 | 3.13 |
ENSMUST00000022507.13
ENSMUST00000163924.2 |
Pspc1
|
paraspeckle protein 1 |
| chrX_+_72818003 | 3.12 |
ENSMUST00000002081.6
|
Srpk3
|
serine/arginine-rich protein specific kinase 3 |
| chr13_-_73848807 | 3.10 |
ENSMUST00000022048.6
|
Slc6a19
|
solute carrier family 6 (neurotransmitter transporter), member 19 |
| chr15_+_6673167 | 3.06 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr11_+_120489358 | 2.98 |
ENSMUST00000093140.5
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr2_+_128942900 | 2.93 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr6_-_67512768 | 2.91 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr8_-_84467798 | 2.88 |
ENSMUST00000075843.13
ENSMUST00000109802.3 ENSMUST00000166939.8 ENSMUST00000002964.14 |
Adgre5
|
adhesion G protein-coupled receptor E5 |
| chrX_+_158623460 | 2.87 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_-_34089109 | 2.86 |
ENSMUST00000085585.12
|
Lsm14a
|
LSM14A mRNA processing body assembly factor |
| chr13_-_23806530 | 2.82 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr6_-_4747066 | 2.81 |
ENSMUST00000090686.11
ENSMUST00000133306.8 |
Sgce
|
sarcoglycan, epsilon |
| chr14_-_51045182 | 2.81 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr4_+_133097013 | 2.81 |
ENSMUST00000030669.8
|
Slc9a1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1 |
| chr6_+_4747356 | 2.78 |
ENSMUST00000176551.3
|
Peg10
|
paternally expressed 10 |
| chr15_-_101389384 | 2.76 |
ENSMUST00000023718.9
|
Krt83
|
keratin 83 |
| chr2_+_128942919 | 2.74 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr11_-_120489172 | 2.71 |
ENSMUST00000026125.3
|
Alyref
|
Aly/REF export factor |
| chr13_-_23755374 | 2.69 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr6_+_5390386 | 2.62 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr12_+_85520652 | 2.60 |
ENSMUST00000021674.7
|
Fos
|
FBJ osteosarcoma oncogene |
| chr3_+_103739877 | 2.59 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr15_-_101602734 | 2.58 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr15_+_91722524 | 2.57 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr11_+_6510167 | 2.55 |
ENSMUST00000109722.9
|
Ccm2
|
cerebral cavernous malformation 2 |
| chr13_+_23936250 | 2.54 |
ENSMUST00000091703.3
|
H3c2
|
H3 clustered histone 2 |
| chr18_-_35855383 | 2.47 |
ENSMUST00000133064.8
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr16_-_92196954 | 2.38 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr8_-_85696369 | 2.34 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr11_+_62441992 | 2.31 |
ENSMUST00000019649.4
|
Ubb
|
ubiquitin B |
| chr7_-_30312246 | 2.27 |
ENSMUST00000006476.6
|
Upk1a
|
uroplakin 1A |
| chr18_-_34757653 | 2.27 |
ENSMUST00000003876.10
ENSMUST00000115766.8 ENSMUST00000097626.10 ENSMUST00000115765.2 |
Brd8
|
bromodomain containing 8 |
| chr7_-_140846328 | 2.26 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr7_-_141241632 | 2.26 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr19_+_60744385 | 2.25 |
ENSMUST00000088237.6
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr19_-_40982576 | 2.23 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr15_-_101621332 | 2.22 |
ENSMUST00000023709.7
|
Krt5
|
keratin 5 |
| chr17_+_29587930 | 2.12 |
ENSMUST00000137644.3
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr9_-_70564403 | 2.10 |
ENSMUST00000213380.2
ENSMUST00000049031.6 |
Mindy2
|
MINDY lysine 48 deubiquitinase 2 |
| chr6_-_78355834 | 2.08 |
ENSMUST00000089667.8
ENSMUST00000167492.4 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr8_+_124138163 | 2.08 |
ENSMUST00000071134.4
ENSMUST00000212743.2 |
Tubb3
|
tubulin, beta 3 class III |
| chr2_-_93787441 | 2.07 |
ENSMUST00000099689.5
|
Gm13889
|
predicted gene 13889 |
| chr13_+_23759930 | 2.05 |
ENSMUST00000105105.4
|
H3c4
|
H3 clustered histone 4 |
| chr17_+_34174782 | 2.05 |
ENSMUST00000025178.17
|
Vps52
|
VPS52 GARP complex subunit |
| chr14_-_50479161 | 1.98 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr17_-_14914484 | 1.93 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr15_-_98380567 | 1.92 |
ENSMUST00000023726.5
|
Lalba
|
lactalbumin, alpha |
| chr1_-_36312482 | 1.92 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr10_+_87926932 | 1.91 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr4_+_150572847 | 1.86 |
ENSMUST00000105680.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr3_-_92393193 | 1.84 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr10_-_79744726 | 1.83 |
ENSMUST00000165684.8
ENSMUST00000164705.8 ENSMUST00000105378.9 ENSMUST00000170409.2 |
Med16
|
mediator complex subunit 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.5 | 43.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 12.4 | 62.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 11.2 | 33.6 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 7.5 | 37.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 7.2 | 21.5 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 5.0 | 59.7 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 4.6 | 13.9 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 4.4 | 17.6 | GO:0036233 | proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 4.2 | 12.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 3.8 | 11.3 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 3.5 | 21.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 3.2 | 6.3 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 3.1 | 27.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 2.9 | 14.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 2.7 | 13.6 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 2.7 | 21.4 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 2.5 | 10.0 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 2.4 | 19.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 2.2 | 6.7 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 2.2 | 17.8 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) regulation of chaperone-mediated autophagy(GO:1904714) |
| 2.1 | 20.6 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 2.0 | 49.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 1.5 | 4.4 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.5 | 8.7 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.4 | 5.7 | GO:0017126 | nucleologenesis(GO:0017126) |
| 1.4 | 4.2 | GO:0090420 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 1.3 | 9.2 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 1.3 | 11.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.3 | 29.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 1.2 | 3.7 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 1.1 | 4.6 | GO:1901079 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) regulation of dense core granule biogenesis(GO:2000705) |
| 1.1 | 7.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.1 | 24.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 1.1 | 6.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 1.0 | 1.9 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.0 | 7.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 1.0 | 3.8 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.9 | 4.7 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.9 | 2.8 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.9 | 6.0 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.8 | 26.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.8 | 8.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.8 | 7.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.8 | 6.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.8 | 6.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.8 | 8.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.7 | 2.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.7 | 4.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.7 | 5.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.7 | 7.2 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.6 | 11.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.6 | 3.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.6 | 4.9 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.6 | 2.9 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.6 | 9.9 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.5 | 7.0 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.5 | 4.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.5 | 5.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 5.6 | GO:0032275 | luteinizing hormone secretion(GO:0032275) |
| 0.5 | 1.5 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.5 | 1.9 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.5 | 1.4 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.5 | 76.4 | GO:0007586 | digestion(GO:0007586) |
| 0.5 | 1.9 | GO:0021699 | cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.5 | 9.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.4 | 4.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.4 | 12.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.4 | 8.8 | GO:0060213 | dentate gyrus development(GO:0021542) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.4 | 12.7 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.4 | 7.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.4 | 1.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.4 | 2.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.4 | 1.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.4 | 12.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 7.0 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.3 | 7.2 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.3 | 15.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.3 | 3.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.3 | 4.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 0.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.3 | 12.2 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.3 | 1.0 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.3 | 3.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 2.6 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 5.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 8.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.2 | 2.8 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.2 | 2.6 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.2 | 1.0 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 14.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 0.7 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 22.3 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.2 | 1.0 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) regulation of ovarian follicle development(GO:2000354) |
| 0.2 | 0.3 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 5.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.3 | GO:1902623 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) negative regulation of monocyte chemotaxis(GO:0090027) regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 2.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.4 | GO:1901561 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.1 | 1.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 2.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.8 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.1 | 8.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 1.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 3.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 2.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.5 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 | 0.3 | GO:2000422 | T-helper 1 cell lineage commitment(GO:0002296) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 0.5 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 2.4 | GO:0007614 | short-term memory(GO:0007614) locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 1.1 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.1 | 0.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 3.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 2.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 9.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.7 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 1.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 3.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 3.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 2.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.7 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 7.9 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 2.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 2.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 4.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 3.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 3.1 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 1.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 6.9 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 2.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 2.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.3 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 2.8 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 2.5 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.6 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.2 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 2.9 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 3.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 11.0 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 0.4 | GO:0021591 | ventricular system development(GO:0021591) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 41.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 4.1 | 57.0 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 2.9 | 11.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.6 | 7.9 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 2.6 | 7.7 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 2.2 | 17.8 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 2.0 | 33.6 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.8 | 19.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.4 | 21.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.3 | 3.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.2 | 39.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 1.2 | 71.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 1.1 | 6.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.0 | 4.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.0 | 4.8 | GO:0044393 | microspike(GO:0044393) |
| 0.9 | 7.4 | GO:0001652 | granular component(GO:0001652) |
| 0.9 | 8.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.9 | 9.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.8 | 7.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.8 | 4.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.8 | 4.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.8 | 6.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.8 | 12.8 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.7 | 3.7 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.7 | 6.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.7 | 12.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.6 | 7.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 7.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 2.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.4 | 2.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 4.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 5.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 8.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 8.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 13.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.3 | 24.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 1.5 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 23.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 5.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 20.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 4.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 1.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 2.8 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 0.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 11.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 5.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 52.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 2.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.0 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 65.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 9.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 24.3 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 8.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 5.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 16.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 10.7 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.7 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 16.0 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 14.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 30.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 3.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.8 | 59.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 11.3 | 11.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 9.5 | 57.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 5.9 | 17.6 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 5.4 | 26.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 5.4 | 42.9 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 4.0 | 59.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 3.4 | 13.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 3.3 | 10.0 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 3.3 | 39.2 | GO:0008061 | chitin binding(GO:0008061) |
| 3.2 | 19.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 3.0 | 21.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 2.5 | 7.6 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.9 | 5.6 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 1.8 | 12.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 1.8 | 8.8 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 1.3 | 24.2 | GO:0016918 | retinal binding(GO:0016918) |
| 1.2 | 8.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.9 | 3.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.9 | 6.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.9 | 4.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.8 | 6.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.8 | 31.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.8 | 23.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 1.5 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.7 | 38.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.7 | 37.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.7 | 6.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.7 | 34.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.6 | 1.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.6 | 3.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 3.8 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.6 | 4.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.6 | 126.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.6 | 4.6 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.6 | 7.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.6 | 3.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.6 | 8.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.5 | 22.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.5 | 2.1 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.5 | 2.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 9.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.5 | 7.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 7.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 8.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.4 | 4.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.4 | 27.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 5.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 2.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 1.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 8.6 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.3 | 2.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 48.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 3.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 6.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 1.8 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 7.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.2 | 24.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 5.9 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.2 | 15.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 5.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 12.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 0.7 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 2.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 5.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 23.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 0.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 8.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 6.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.9 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 18.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 4.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 4.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 7.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 6.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.3 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 16.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 3.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 3.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 8.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 5.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 2.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 6.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.6 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 2.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 50.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.7 | 57.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.6 | 33.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 17.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.5 | 17.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 44.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.4 | 9.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 82.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 7.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.3 | 33.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 15.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 15.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 6.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 7.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 17.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 9.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 11.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 8.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 3.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 3.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 57.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.6 | 26.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.3 | 39.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.1 | 71.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.8 | 11.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.8 | 24.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.7 | 15.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.6 | 41.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 21.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.6 | 5.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.5 | 8.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 6.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 7.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 15.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 17.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 4.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 20.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 5.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 11.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 4.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 2.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 5.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 11.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 14.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 6.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 2.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 2.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 4.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 8.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 35.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 3.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 6.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 3.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |